Mutations
APOE
APOE encodes a secreted apolipoprotein involved in lipid metabolism. It is best known for its three major alleles—APOE2, APOE3, and APOE4 —with APOE4 increasing the risk for Alzheimer’s disease and APOE2 reducing it. Every person has an APOE genotype determined by pairs of these alleles: APOE3/E3 (the most common), APOE3/E4, APOE3/E2, APOE2/E4, APOE4/E4, or APOE2/E2. Other variants , many of them rare, have also been studied. Some have been tied to AD, to other neurological disorders, and/or to peripheral conditions, including alterations of lipid metabolism, cardiovascular or kidney disease.
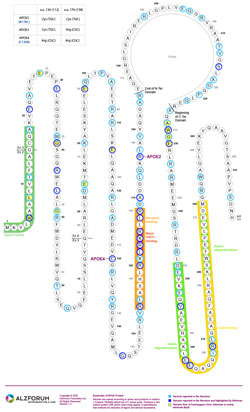
This dataset uses the sequence of the most common APOE allele, APOE3, as a reference. Departures from this sequence are listed as variants. Amino acid position is based on the full-length ApoE protein, 317 amino acids long, including the signal peptide. Positions based on the mature protein, 299 amino acids long, appear in gray (diagram) and in parentheses (table). References for ApoE domains and regions can be found in Methods.
Click on the amino acids in the diagram or on the “Additional Variants” box in the lower-right-hand corner. You can also click on the variants in the table below. See Dec 2022 news for more information.
Search Results
APOE (154)
To request a download of the data in this table, please email mutations@alzforum.org
| Mutation | Clinical Phenotype Studied |
DNA Change | Expected RNA | Protein Consequence | Coding/Non-Coding | Genomic Region | Biological Effect | Primary Papers |
|---|---|---|---|---|---|---|---|
| APOE Region
|
Alzheimer's Disease, Multiple Conditions | Both | Multiple effects. |
||||
| c.-1334G>A
(rs7259620) |
Alzheimer's Disease, Blood Lipids/Lipoproteins, Cardiovascular Disease | Substitution | Non-Coding | 2kb upstream | Unknown, but predicted benign in silico (PHRED-scaled CADD =0.15). |
Takei et al., 2009 | |
| c.-779_-776del
(rs538385866) |
Alzheimer's Disease | Deletion | Non-Coding | 2kb upstream | Unknown, but located in predicted promoter region. |
Yee et al., 2021 | |
| c.-647A>G
(rs439382) |
Alzheimer's Disease | Substitution | Non-Coding | 2kb upstream | Unknown, but predicted benign in silico (PHRED-scaled CADD =0.45). |
Reitz et al., 2013 | |
| g.45408560_45410359del
(DEL 19:44905303-44907102) |
Alzheimer's Disease | Deletion | Deletion | Deletion | Both | Promoter, Exon 1, Exon 2 | Predicted to eliminate APOE expression. |
Chemparathy et al., 2024 |
| c.-558A>T
(rs449647, -491A/T) |
Alzheimer's Disease, Multiple Conditions | Substitution | Non-Coding | 2kb upstream | Unclear, but several studies reported effects on APOE transcription. |
Bullido et al., 1998 | |
| c.-494T>C
(rs769446, -427 T/C, -427C/T) |
Alzheimer's Disease, Multiple Conditions | Substitution | Non-Coding | 2kb upstream | Unclear, but some studies reported effects on APOE transcription. |
Artiga et al., 1998; Artiga et al., 1998 |
|
| c.-488C>A
(rs532314089) |
Alzheimer's Disease | Substitution | Non-Coding | 2kb upstream | Unknown, but predicted to disrupt binding of transcription factor EGR1. PHRED-scaled CADD = 0.26. |
Yee et al., 2021 | |
| c.-380A>G
(rs1038445539) |
Hyperlipoproteinemia Type IIb | Substitution | Non-Coding | 2kb upstream | Unknown, but predicted benign in silico (PHRED-scaled CADD = 7). |
Abou Khalil et al., 2022 | |
| c.-286T>G
(rs405509, -219T/G, -219G/T, Th1/E47cs) |
Alzheimer's Disease, Multiple Conditions | Substitution | Non-Coding | 2kb upstream | Modified APOE transcription. |
Artiga et al., 1998; Lambert et al., 1998 |
|
| c.-279G>A
|
Hyperlipoproteinemia Type IIb | Substitution | Non-Coding | 2kb upstream | Unknown, but predicted benign in silico (PHRED-scaled CADD = 6). |
Abou Khalil et al., 2022 | |
| c.-233G>C
|
Hyperlipoproteinemia Type IIa | Substitution | Non-Coding | 2kb upstream | Unknown, but predicted benign in silico (PHRED-scaled CADD = 10). |
Abou Khalil et al., 2022 | |
| c.-105A>G
|
Hyperlipoproteinemia Type IIb | Substitution | Non-Coding | 2kb upstream | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 23). |
Abou Khalil et al., 2022 | |
| c.-81G>A
(rs766215051) |
Hyperlipoproteinemia Type IIa | Substitution | Substitution | | Non-Coding | Exon 1, 5' UTR | Unknown, but its PHRED-scaled CADD score (14) did not reach the commonly used threshold of 20 for predicting deleteriousness. |
Abou Khalil et al., 2022 |
| c.-78C>G
(rs750782549) |
Hyperlipoproteinemia Type IIa, Hyperlipoproteinemia Type IIb | Substitution | Substitution | | Non-Coding | Exon 1, 5' UTR | Unknown, but its PHRED-scaled CADD score (15) did not reach the commonly used threshold of 20 for predicting deleteriousness. |
Abou Khalil et al., 2022 |
| c.-24+38G>A
(rs373985746) |
Alzheimer's Disease | Substitution | Substitution | | Non-Coding | Intron 1 | Unknown, but predicted benign in silico (PHRED-scaled CADD =10). |
Yee et al., 2021 |
| c.-24+69C>G
(rs440446, +113C/G, IE1, IVS1+69, +969) |
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | | Non-Coding | Intron 1 | Modified APOE transcription. |
Mui et al., 1996 |
| c.-24+288G>A
(rs192348494) |
Alzheimer's Disease | Substitution | Substitution | | Non-Coding | Intron 1 | Unknown, but predicted benign in silico (PHRED-scaled CADD =12). |
Yee et al., 2021 |
| c.-23-377A>G
(rs150375400) |
Alzheimer's Disease | Substitution | Substitution | | Non-Coding | Intron 1 | Unknown, but predicted benign in silico (PHRED-scaled CADD =10). |
Yee et al., 2021 |
| c.-23-280C>T
(rs769448) |
Blood Lipids/Lipoproteins | Substitution | Substitution | | Non-Coding | Intron 1 | Unknown, but predicted to modify APOE transcription. |
Radwan et al., 2014 |
| W5Ter
(p.W5*, p.Thr5*) |
Alzheimer's Disease, Blood Lipids/Lipoproteins | Substitution | Substitution | Nonsense | Coding | Exon 2 | Predicted to abrogate production of ApoE as it introduces a stop codon in the signal peptide. PHRED-scaled CADD = 35. |
Leren et al., 2016 |
| L8Ter
(p.L8*) |
Alzheimer's Disease | Substitution | Substitution | Nonsense | Coding | Exon 2 | Predicted to eliminate functional ApoE expression. |
Chemparathy et al., 2024 |
| T11S
|
Alzheimer's Disease, Blood Lipids/Lipoproteins, Dementia | Substitution | Substitution | Missense | Coding | Exon 2 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 0.6). |
Rasmussen et al., 2020 |
| T11A
|
Hyperlipoproteinemia Type IIb | Substitution | Substitution | Missense | Coding | Exon 2 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 0.3). |
Abou Khalil et al., 2022 |
| c.43+11G>A
(rs770658351) |
Hyperlipoproteinemia Type IIb | Substitution | Substitution | | Non-Coding | Intron 2 | Unknown, but its PHRED-scaled CADD score (13) did not reach the commonly used threshold of 20 for predicting deleteriousness. |
Abou Khalil et al., 2022 |
| c.43+64C>T
(rs143063029) |
Substitution | Substitution | | Non-Coding | Intron 2 | Unknown, but predicted benign in silico (PHRED-scaled CADD=4.94). |
Chakraborty and Kahali, 2023 | |
| c.43+78G>A
(rs769449) |
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | | Non-Coding | Intron 2 | Unknown, but may modify APOE transcription. |
Ridker et al., 2008 |
| c.43+219_43+221dupGTT
(rs374670655) |
Alzheimer's Disease | Duplication | Duplication | | Non-Coding | Intron 2 | Unknown. |
Yee et al., 2021 |
| c.43+349G>A
(rs61357706, APOE2269) |
Blood Lipids/Lipoproteins | Substitution | Substitution | | Non-Coding | intron 2 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 6). |
Radwan et al., 2014 |
| c.43+520G>A
(rs769450, Int2 G/A) |
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | | Non-Coding | Intron 2 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 4). |
He et al., 2009 |
| c.44-523G>C
(rs1021908335) |
Alzheimer's Disease | Substitution | Substitution | | Non-Coding | Intron 2 | Unknown, but predicted benign in silico (PHRED-scaled CADD =0.7). |
Yee et al., 2021 |
| c.44-469A>G
(rs115299243) |
Blood Lipids/Lipoproteins | Substitution | Substitution | | Non-Coding | Intron 2 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 6). |
Radwan et al., 2014 |
| c.44-1G>C
|
Hyperlipoproteinemia Type IIa | Substitution | Splicing Alteration | Deletion | Non-Coding | Intron 2 | Unknown, but predicted to alter splicing; predicted damaging in silico (PHRED-scaled CADD = 33). |
Abou Khalil et al., 2022 |
| A18T
|
Alzheimer's Disease, Cardiovascular Disease | Substitution | Substitution | Missense | Coding | Exon 3 | Predicted to disrupt signal peptide cleavage and affect ApoE secretion. PHRED-scaled CADD = 22. |
Zhou et al., 2018; Yee et al., 2021 |
| E21K (E3K)
(ApoE5) |
Multiple Conditions | Substitution | Substitution | Missense | Coding | Exon 3 | Increased receptor binding in cell-based LDL competition assay; no effect on heparin binding. |
Yamamura et al., 1984; Tajima et al., 1988 |
| A23V (A5V)
|
Alzheimer's Disease, Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 3 | Decreased plasma ApoE levels. |
Rasmussen et al., 2020 |
| A23A (A5A)
|
Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type IIb | Substitution | Substitution | Silent | Coding | Exon 3 | Unknown, but predicted benign in silico (PHRED-scaled 3.7). |
Radwan et al., 2014 |
| E27fs (E9fs)
|
Blood Lipids/Lipoproteins, Kidney Disorder: Nephrotic Syndrome | Deletion | Deletion | Frame Shift | Coding | Exon 3 | Unknown, predicted to be degraded by nonsense-mediated mRNA decay. |
Corredor-Andrés et al., 2018 |
| E31K (E13K)
(ApoE5 French-Canadian, ApoE4 Philadelphia) |
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | Missense | Coding | Exon 3 | Unknown. In heparin-binding site, but predicted benign in silico (PHRED-scaled CADD = 12). |
Mailly et al., 1991 |
| W38Ter (W20Ter)
|
Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type III | Substitution | Substitution | Nonsense | Coding | Exon 3 | Predicted to produce a truncated protein and/or low levels of ApoE, but ApoE levels in carriers were not consistently lower; PHRED-scaled CADD = 29. |
Feussner et al., 1998 |
| Q39Ter (Q21Ter)
(p.Q39*) |
Alzheimer's Disease | Substitution | Substitution | Nonsense | Coding | Exon 3 | Predicted to eliminate functional APOE expression. |
Chemparathy et al., 2024 |
| R43C (R25C)
(Kyoto, ApoE2 Kyoto) |
Multiple Conditions | Substitution | Substitution | Missense | Coding | Exon 3 | Altered receptor binding, increased aggregation. |
Matsunaga et al., 1999 |
| L46P (L28P)
(ApoE4 Freiburg, ApoE4 Pittsburgh) |
Alzheimer's Disease, Hyperlipoproteinemia Type IIa, Hyperlipoproteinemia Type IIb | Substitution | Substitution | Missense | Coding | Exon 3 | Increased Aβ uptake in neurons, increased reactive oxygen species, reduced cell viability. Altered HDL metabolism. Unstable, abnormal structure, more prone to proteolysis. |
Kamboh et al., 1999; Orth et al., 1999 |
| G49fs (G31fs)
|
Hyperlipoproteinemia Type III | Deletion | Deletion | Frame Shift | Coding | Exon 3 | Eliminates protein, likely by nonsense-mediated decay of mRNA. |
Feussner et al., 1992 |
| R50C (R32C)
|
Substitution | Substitution | Missense | Coding | Exon 3 | Unknown, but PHRED-scaled CADD score predicted deleterious (25.3). |
Pires et al., 2017 | |
| R50H (R32H)
|
Cardiovascular Disease, Kidney Disorder: Lipoprotein Glomerulopathy | Substitution | Substitution | Missense | Coding | Exon 3 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 23). |
Li et al., 2022 |
| T60A (T42A)
(ApoE3 Freiburg) |
Alzheimer's Disease, Blood Lipids/Lipoproteins, Cardiovascular Disease | Substitution | Substitution | Missense | Coding | Exon 3 | Unknown, did not reach deleterious threshold in silico (PHRED-scaled CADD = 15). |
Wieland et al., |
| Q64H (Q46H)
|
Alzheimer's Disease | Substitution | Substitution | Missense | Coding | Exon 3 | Unknown, but predicted damaging (PHRED-scaled CADD = 23.3). |
Yee et al., 2021 |
| c.236+50T>C
(rs12982192) |
Alzheimer's Disease | Substitution | Splicing Alteration | | Non-Coding | Intron 3 | Modified APOE transcription. |
Dieter and Estus, 2010 |
| c.237-33C>G
|
Hyperlipoproteinemia Type III | Substitution | Substitution | | Non-Coding | Intron 3 | Unknown, but in silico algorithms predicted it is non-deleterious (PHRED-scaled CADD = 11.42). |
Bea et al., 2023 |
| c.237-28C>T
(rs372675300) |
Alzheimer's Disease | Substitution | Substitution | | Non-Coding | Intron 3 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 2.79). |
Yee et al., 2021 |
| c.237-1A>G
(3592 A>G) |
Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type III | Substitution | Splicing Alteration | | Non-Coding | intron 3 | Disrupted splicing resulting in near abrogation of ApoE expression. |
Ghiselli et al., 1981; Cladaras et al., 1987 |
| D83D (D65D)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 0.6). |
Abou Khalil et al., 2022 |
| E84Ter (E66Ter)
(p.E84*) |
Progressive Supranuclear Palsy | Substitution | Substitution | Nonsense | Coding | Exon 4 | Predicted to abrogate ApoE full-length synthesis introducing an early stop codon. |
Chemparathy et al., 2024 |
| K90E (K72E)
|
Hyperlipoproteinemia Type IIb | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 22). |
Evans et al., 2013 |
| E98fs (E80fs)
(E97fs) |
Alzheimer's Disease, Cardiovascular Disease, Hyperlipoproteinemia Type III | Deletion | Deletion | Frame Shift | Coding | Exon 4 | Eliminates expression of ApoE protein. |
Mak et al., 2014 |
| Q99K (Q81K)
(ApoE5 Frankfurt) |
Blood Lipids/Lipoproteins, Cardiovascular Disease | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but in silico prediction did not reach commonly used threshold for deleteriousness (PHRED-scaled CADD = 19). |
Ruzicka et al., 1993 |
| P102T (P84T)
|
Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but PHRED-scaled CADD score predicted deleterious (22.5). |
Pires et al., 2017 | |
| P102R (P84R)
(ApoE5) |
Blood Lipids/Lipoproteins, Cardiovascular Disease | Substitution | Substitution | Missense | Coding | Exon 4 | No effect on receptor binding detected, but predicted deleterious by wide variety of in silico algorithms. |
Wardell et al., 1991 |
| P102L (P84L)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 23). |
Abou Khalil et al., 2022 |
| A104A (A86A)
(A104=, A86=) |
Hyperlipoproteinemia Type IV | Substitution | Substitution | Silent | Coding | Exon 4 | Unknown, but in silico algorithms predicted it is non-deleterious (PHRED-scaled CADD = 1.10). |
Bea et al., 2023 |
| R108W (R90W)
|
Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown. In silico predictions were inconsistent, but PHRED-scaled CADD score (23.6) suggests deleteriousness. |
Bea et al., 2023 |
| R110P (R92P)
|
Blood Lipids/Lipoproteins, Pancreatitis | Substitution | Substitution | Missense | Coding | Exon 4 | Unkown, but in silico algorithm predicted deleterious (PHRED-CADD = 24.6). |
Abedi et al., 2023 |
| E114fs (E96fs)
(ApoE3 Groningen) |
Hyperlipoproteinemia Type III | Insertion | Insertion | Frame Shift | Coding | Exon 4 | Predicted to result in a truncated ApoE species or in elimination of its production. |
Dijck-Brouwer et al., 2005 |
| E114K (E96K)
|
Alzheimer's Disease, Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but carriers had reduced ApoE levels in plasma. |
Rasmussen et al., 2020 |
| R121W (R103W)
|
Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious (PHRED-scaled CADD = 26.1). |
Bea et al., 2023 |
| A124V (A106V)
(ApoE3 Basel) |
Alzheimer's Disease, Blood Lipids/Lipoproteins, Cardiovascular Disease | Substitution | Substitution | Missense | Coding | Exon 4 | No effect on receptor binding, internalization, or degradation. |
Miserez et al., 2003 |
| C130R (C112R)
(ApoE4) |
Alzheimer's Disease, Multiple Conditions | Allele | Allele | Allele | Coding | Exon 4 | Strongest known risk factor for AD. Implicated in Aβ and tau pathologies, as well as in multiple other neuronal and non-neuronal functions. |
Weisgraber et al., 1981; Zannis and Breslow, 1981 |
| [C130R;R176C] ([C112R;R158C])
(ApoE3r, ApoE1y) |
Blood Lipids/Lipoproteins, Motor Neuron Disease | Coding | Exon 4 | Unknown. |
Persico et al., 2004 | ||
| R132C (R114C)
(Tsukuba) |
Splenomegaly | Substitution | Substitution | Missense | Coding | Exon 4 | Enhanced aggregation, destabilized protein. |
Hagiwara et al., 2008 |
| R132S (R114S)
|
Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico by multiple algorithms. |
Pires et al., 2017 | |
| R132P (R114P)
|
Cardiovascular Disease, Diabetes Mellitus | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 25). |
Stephens et al., 2005 |
| Y136H (Y118H)
|
Blood Lipids/Lipoproteins | Coding | Exon 4 | Unknown, but in silico algorithms predicted it is deleterious (PHRED-scaled CADD = 24.8) |
Bea et al., 2023 | ||
| R137C (R119C)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 26). |
Abou Khalil et al., 2022 |
| R137H (R119H)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 22). |
Abou Khalil et al., 2022 |
| G145D (G127D)
(ApoE1 Bethesda) |
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | Missense | Coding | Exon 4 | Decreased heparin binding and altered b-VLDL binding (LRP receptor), with no apparent effect on LDL binding. |
Weisgraber et al., 1984; Gregg et al., 1983 |
| G145InsEVQAMLG (G127InsEVQAMLG)
(Leiden) |
Hyperlipoproteinemia Type III | Duplication | Duplication | Duplication | Coding | Exon 4 | Reduced receptor binding, heparin binding, HSPG-LPL binding, and lipolysis; increased preference for VLDL. |
Wardell et al., 1989; van den Maagdenberg et al., 1989; Havekes et al., 1986 |
| R152W (R134W)
|
Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but PHRED-scaled CADD score predicted deleterious (23.3). |
Pires et al., 2017 | |
| R152Q (R134Q)
|
Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 23). |
de Knijff et al., 1994 | |
| R154S (R136S)
(Christchurch, ApoE2-Christchurch, APOE3 Christchurch) |
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced receptor and HSPG binding; reduced amyloid-β aggregation, tau phosphoryation. Increased tau binding and decreased tau proteolysis and uptake by neurons and microglia. In mouse models of AD and tauopathy, reduced Aβ-induced tau seeding and spreading. In APOE4 context, reduced tauopathy, neurodegeneration, and neuroinflammation in mice and iPSCs. |
Wardell et al., 1987; Emi et al., 1988 |
| R154C (R136C)
|
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced receptor binding. |
Walden et al., 1994 |
| R154fs (R136fs)
(ApoE Australia, ApoE0) |
Hyperlipoproteinemia Type III | Deletion | Deletion | Frame Shift | Coding | Exon 4 | Absence of ApoE, at least in VLDL lipoprotein. |
Tate et al., 2001 |
| R154H (R136H)
|
Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Moderately reduced receptor and heparin binding. |
Minnich et al., 1995 |
| L155F (L137F)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 6). |
Abou Khalil et al., 2022 |
| K161_R165del (K143_R147del)
|
Blood Lipids/Lipoproteins, Kidney Disorder: Lipoprotein Glomerulopathy | Deletion | Deletion | Deletion | Coding | Exon 4 | Unknown, but expected to alter receptor binding and heparin binding. |
Xie et al., 2019 |
| R160S (R142S)
|
Hyperlipoproteinemia Type IIa, Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 26). |
Sakuma et al., 1995; Sakuma et al., 2014 |
| R160C (R142C)
(ApoE1 Nagoya) |
Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced receptor binding and especially heparin binding. Increased prevalence in VLDLs. Multiple effects on lipid and lipoprotein profiles in mice. |
Rall et al., 1989; Havel et al., 1983 |
| R160L (R142L)
|
Cardiovascular Disease, Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 28). |
Richard et al., 1995; Richard et al., 1994 |
| V153_R160dup (V135_R142dup)
(ApoE5ss) |
Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type IV | Duplication | Duplication | Duplication | Coding | Exon 4 | Unknown, but alters the receptor binding and main heparin binding regions. |
Yamanouchi et al., 2001 |
| L162_K164del (L144_K146del)
(Tokyo/Maebashi, ApoE Tokyo, ApoE Maebashi, L141_K143del, R142_L144del, K143_R145del) |
Blood Lipids/Lipoproteins, Kidney Disorder: Lipoprotein Glomerulopathy | Deletion | Deletion | Deletion | Coding | Exon 4 | Unknown, but expected to alter receptor binding and heparin binding. |
Konishi et al., 1999; Ogawa et al., 2000; Maruyama et al., 1997 |
| R163C (R145C)
(ApoE4 Philadelphia, ApoE Qatar, APOE ε3[R145C]) |
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced heparin binding and moderately reduced receptor binding. |
Rall et al., 1982 |
| R163H (R145H)
(ApoE Kochi) |
Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 23). |
Suehiro et al., 1990 |
| R163P (R145P)
(Sendai) |
Kidney Disorder: Lipoprotein Glomerulopathy | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced receptor and heparin binding in vitro. In mice, caused LPG-like renal aggregates, altered lipid/lipoprotein profile in blood, and altered macrophage function. |
Oikawa et al., 1991; Oikawa et al., 1997 |
| K164Q (K146Q)
|
Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Altered receptor and heparin binding; reduced VLDL lipolysis. |
Rall et al., 1983; Smit et al., 1990 |
| K164E (K146E)
(ApoE1 Harrisburg) |
Blood Lipids/Lipoproteins, Cardiovascular Disease, Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced receptor and heparin binding, and reduced ApoE turnover. May interfere with the coordination between lipid association and receptor binding. |
Mann et al., 1989; Mann et al., 1995 |
| K164_R165delinsNW (K146_R147delinsNW)
(ApoE1 Hammersmith) |
Hyperlipoproteinemia Type III | Deletion-Insertion | Deletion-Insertion | Deletion-Insertion | Coding | Exon 4 | In mice, drastically altered lipid and lipoprotein profiles in blood. May act as dominant-negative receptor ligand and interfere with LPL and LCAT activities. |
Hoffer et al., 1996 |
| R165W
|
Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced binding to LDL receptors. |
Bea et al., 2023 |
| R165P (R147P)
(Chicago) |
Kidney Disorder: Lipoprotein Glomerulopathy | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but showed enhanced binding to glomerular capillaries. Also, induced structural changes resulting in reduced ApoE stability. Predicted to disrupt oligomerization, enhance aggregation, and alter receptor and heparin binding. |
Sam et al., 2006 |
| L167del (L149del)
|
Multiple Conditions | Deletion | Deletion | Deletion | Coding | Exon 4 | Altered receptor binding, possibly increasing binding affinity. Also altered lipoprotein catabolism, and possibly lipolytic activity. |
Nguyen et al., 2000 |
| R168C (R150C)
(ApoE Modena) |
Blood Lipids/Lipoproteins, Kidney Disorder: Lipoprotein Glomerulopathy, Splenomegaly | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but may induce dimer formation and facilitate entrapment in kidney capillaries. Also, may affect receptor binding. |
Russi et al., 2009 |
| R168G (R150G)
(Okayama) |
Blood Lipids/Lipoproteins, Kidney Disorder: Lipoprotein Glomerulopathy | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 23). |
Kinomura et al., 2008 |
| R168H (R150H)
|
Alzheimer's Disease, Blood Lipids/Lipoproteins, Diabetes Mellitus | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 23). |
Stephens et al., 2005 |
| R168P (R150P)
(ApoE Guangzhou) |
Blood Lipids/Lipoproteins, Kidney Disorder: Lipoprotein Glomerulopathy | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 23). |
Luo et al., 2008 |
| D169dup (D151dup)
(ApoE2 Kanto) |
Kidney Disorder: Lipoprotein Glomerulopathy | Duplication | Duplication | Duplication | Coding | Exon 4 | Unknown. |
Yokochi et al., 2024; Matsunaga and Saito, 2014 |
| A170P (A152P)
|
Substitution | Substitution | Missense | Coding | Exon 4 | Reduced receptor binding. |
McLean et al., 1984 | |
| A170D (A152D)
(ApoE Las Vegas) |
Blood Lipids/Lipoproteins, Kidney Disorder: Lipoprotein Glomerulopathy | Substitution | Substitution | Missense | Coding | Exon 4 | Aggregation-prone. Reduced helical content, stability, predicted to induce misfolding. |
Bomback et al., 2010 |
| L173L (L155L)
|
Hyperlipoproteinemia Type IIb | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 8). |
Abou Khalil et al., 2022 |
| L173P (L155P)
(ApoE Chengdu) |
Kidney Disorder: Lipoprotein Glomerulopathy, Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 25). |
Wu et al., 2018 |
| Q174_G191del (Q156_G173del)
|
Kidney Disorder: Lipoprotein Glomerulopathy, Cardiovascular Disease, Blood Lipids/Lipoproteins | Deletion | Deletion | Deletion | Coding | Exon 4 | Unknown. |
Ando et al., 1999 |
| R176C (R158C)
(ApoE2) |
Alzheimer's Disease, Multiple Conditions | Allele | Allele | Allele | Coding | Exon 4 | Protective against AD risk. Effects on both Aβ (direct) and tau (possibly indirect) pathologies. Also implicated in multiple neuronal and non-neuronal functions. |
Weisgraber et al., 1981; Zannis and Breslow, 1981 |
| [R176C];[C130R] ([R158C;C112R])
(ApoE2/4, ApoE4/2) |
Alzheimer's Disease, Multiple Conditions | Coding | Exon 4 | When present in the same host, APOE2 appears to partially mitigate APOE4's harmful effects. |
Farrer et al., 1997 | ||
| R176P (R158P)
(Osaka/Kurashiki, ApoE Osaka/Kurashiki, ApoE2 Kurashiki, ApoE Osaka) |
Blood Lipids/Lipoproteins, Cardiovascular Disease, Kidney Disorder: Lipoprotein Glomerulopathy | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but structural analyses predicted changes affecting lipid and receptor binding. |
Tokura et al., 2011; Mitani et al., 2011 |
| L177P (L159P)
(ApoE3 Seongnam) |
Alzheimer's Disease | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 24) with structual consequences. |
Bagaria et al., 2022 |
| V179A (V161A)
|
Hyperlipoproteinemia Type IIb | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 24). |
Abou Khalil et al., 2022 |
| A184fs (A166fs)
(166delG) |
Diabetes Mellitus | Deletion | Deletion | Frame Shift | Coding | Exon 4 | Unknown, but reduced plasma ApoE levels. |
Stephens et al., 2005 |
| A184D (A166D)
|
Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 23). |
Kim et al., 2020 |
| R185R (R167R)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 7). |
Abou Khalil et al., 2022 |
| G191C (G173C)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced affinity for LDL receptors in vitro. |
Bea et al., 2023 |
| R198C (R180C)
(ApoE1 Baden) |
Hyperlipoproteinemia Type IV, Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 26). |
Hoffmann et al., 2001 |
| E204Ter (R186Ter)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Nonsense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 37). |
Sánchez et al., 2021 |
| Q205E (Q187E)
(ApoE2 Toranomon) |
Hyperlipoproteinemia Type III, Cardiovascular Disease, Kidney Disorder | Substitution | Substitution | Missense | Coding | Exon 4 | Receptor and heparin binding appeared to be similar to those of wildtype ApoE3 protein. |
Okubo et al., 1998 |
| R207C (R189C)
|
Cardiovascular Disease | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted deleterious in silico (PHRED-scaled CADD = 25). |
Zhou et al., 2018 |
| T212S (T194S)
|
Hyperlipoproteinemia Type III | | Missense | Coding | Exon 4 | Unknown, but predicted benign in silico. May affect binding of ApoE to Ab. |
Evans et al., 2013 | |
| V213E (V195E)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but its PHRED-scaled CADD score (11) did not reach the commonly used threshold of 20 for predicting deleteriousness. |
Abou Khalil et al., 2022 |
| S215C (S197C)
(Toyonaka) |
Kidney Disorder, Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but its PHRED-scaled CADD score was just below the commonly used threshold of 20 to predict deleteriousness (PHRED-scaled CADD = 19.5). |
Fukunaga et al., 2017 |
| A217A (A199A)
|
Hyperlipoproteinemia Type IIa, Hyperlipoproteinemia Type IIb, Hyperlipoproteinemia Type III | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 7.2). |
Abou Khalil et al., 2022 |
| G218C (G200C)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 7). |
Abou Khalil et al., 2022 |
| P220L (P202L)
|
Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type IIb | Coding | Exon 4 | Increased affinity for LDL receptors in vitro. |
Bea et al., 2023 | ||
| Q222K (Q204K)
|
Substitution | Substitution | Missense | Coding | Exon 4 | Unable to form homodimers and ApoE-ApoAII complexes, but likely due to its ApoE4 backbone. |
Scacchi et al., 2003; Corbo et al., 1999; Corbo et al., 1995 |
|
| A225T (A207T)
|
Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but PHRED-scaled CADD score predicted deleterious (20.8). |
Pires et al., 2017 | |
| A227_E230del (A209_E212del)
|
Blood Lipids/Lipoproteins, Cardiovascular Disease, Hyperlipoproteinemia Type III | Deletion | Deletion | Frame Shift | Coding | Exon 4 | Predicted to cause a frameshift introducing a stop codon at amino acid 247. In homozygous form, resulted in near elimination of ApoE protein. |
Feussner et al., 1996 |
| W228Ter (W210Ter)
(ApoE3 Washington) |
Alzheimer's Disease, Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type III | Substitution | Substitution | Nonsense | Coding | Exon 4 | Generated a truncated protein and, in homozygous form, nearly eliminated ApoE from plasma. In cells, decreased ApoE production and secretion by ~2/3. Lipoprotein and lipid profiles in blood indicated a reduction in hepatic removal of remnant lipoprotein particles. |
Lohse et al., 1992 |
| E230K (E212K)
(ApoE5) |
Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Increased heparin binding, while decreasing cellular internalization and degradation in cultured cells. |
Feussner et al., 1996 |
| R233G (R215G)
|
Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but PHRED-scaled CADD score predicted deleterious (23.7). |
Pires et al., 2017 | |
| R235W (R217W)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 25). |
Marduel et al., 2013 |
| R242Q (R224Q)
(ApoE2 Fukuoka) |
Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Binding to LDL cell surface receptors and heparin was similar to wildytpe ApoE3. |
Moriyama et al., 1996 |
| R246C (R228C)
(ApoE2 Dunedin) |
Hyperlipoproteinemia Type IV, Hyperlipoproteinemia Type V | Substitution | Substitution | Missense | Coding | Exon 4 | Receptor-binding activity was normal in a competitive binding assay. |
Wardell et al., 1990; Nye et al., 1986 |
| D248Y (D230Y)
(ApoE Hong Kong) |
Kidney Disorder: Lipoprotein Glomerulopathy | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 27). |
Cheung et al., 2009 |
| E249K (E231K)
|
Hyperlipoproteinemia Type IIb | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown; its PHRED-scaled CADD score (19.7) was very close to the commonly used threshold of 20 for predicting deleteriousness. |
Abou Khalil et al., 2022 |
| E252K (E234K)
|
Hyperlipoproteinemia Type IIb | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 22). |
Abou Khalil et al., 2022 |
| V254E (V236E)
(Jacksonville, APOE3-Jacksonville, APOE3-Jac, APOE ε3 (V236E), APOE*2(Val236-->Glu)) |
Alzheimer's Disease, Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Reduced ApoE self-oligomerization and promoted lipid metabolism in transgenic mouse brains. Also reported to increase dimerization. In AD mice, reduced plaques and dystrophic neurites. |
van den Maagdenberg et al., 1993 |
| E262_E263delinsKK (E244_E245delinsKK)
(Suita, ApoE7 Suita) |
Multiple Conditions | Deletion-Insertion | Deletion-Insertion | Deletion-Insertion | Coding | Exon 4 | Unclear, but predicted damaging by multiple in silico algorithms. |
Maeda et al., 1989; Yamamura et al., 1984 |
| E262K (E244K)
|
Blood Lipids/Lipoproteins, Cardiovascular Disease | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-CADD = 23.3) |
Maeda et al., 1989; Yamamura et al., 1984 |
| E263K (E245K)
|
Cerebral Palsy, Blood Lipids/Lipoproteins, Cardiovascular Disease | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-CADD = 23.9) |
Maeda et al., 1989; Yamamura et al., 1984 |
| I268M (I250M)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but did not reach commonly used in silico threshold for deleteriousness (PHRED-scaled CADD = 9). |
Sánchez et al., 2021 |
| R269G (R251G)
(ApoE ε4 (R251G)) |
Alzheimer's Disease, Multiple Conditions | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 23). |
van den Maagdenberg et al., 1993 |
| R269H (R251G)
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 25). |
Diboun et al., 2022 |
| L270E (L252E)
|
Hyperlipoproteinemia Type IIa | Deletion-Insertion | Deletion-Insertion | Missense | Coding | Exon 4 | Unknown. |
van den Maagdenberg et al., 1993 |
| R292H (R274H)
|
Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted damaging in silico (PHRED-scaled CADD = 25). |
van den Maagdenberg et al., 1993 |
| W294C (W276C)
|
Substitution | Substitution | Missense | Non-Coding | Exon 4 | Unknown, but predicted to alter ApoE structure by in silico analyses. |
Pires et al., 2017 | |
| S314R (S296R)
|
Blood Lipids/Lipoproteins | Substitution | Substitution | Missense | Coding | Exon 4 | Unknown, but predicted benign in silico (PHRED-scaled CADD = 11). |
van den Maagdenberg et al., 1993 |
| c.*25C > T
(rs374329439) |
Blood Lipids/Lipoproteins, Hyperlipoproteinemia Type IIa, Hyperlipoproteinemia Type IIb | Substitution | Substitution | | Non-Coding | Exon 4, 3' UTR | Unknown, but predicted benign in silico (PHRED-scaled CADD = 6). |
Abou Khalil et al., 2022; Radwan et al., 2014 |
| c.*36C > G
|
Hyperlipoproteinemia Type IIa | Substitution | Substitution | | Non-Coding | Exon 4, 3' UTR | Unknown, but predicted benign in silico (PHRED-scaled CADD = 7). |
Abou Khalil et al., 2022 |
| c.*92C>T
(rs553874843) |
Alzheimer's Disease | Substitution | Substitution | | Non-Coding | Exon 4, 3' UTR | Unknown, but predicted benign in silico (PHRED-scaled CADD = 1.25). |
Yee et al., 2021 |
