Proteomics Highlight Alzheimer’s Changes in Matrisome, MAPK Signaling
Quick Links
In Alzheimer’s disease, proteins in the brain pile up in plaques and tangles. Could the proteome yield clues on how that happens that genomic and transcriptomic studies miss? In today’s Nature Neuroscience, researchers led by Nicholas Seyfried at Emory University School of Medicine, Atlanta, suggest as much. They analyzed more than 8,600 proteins from nearly 1,000 postmortem brain samples, looking for sets of proteins that changed in coordinated ways from control to AD brains. They identified 44 such modules. Two stood out. A set of extracellular matrix, or “matrisome,” proteins rose in tandem with plaques and tangles, with a whopping correlation coefficient of 0.75. Meanwhile, a MAPK signaling module was linked to cognitive decline independently of brain pathology. Curiously, neither module showed up in transcriptome data from the same brains.
- In-depth proteome analysis links extracellular matrix to plaques and tangles.
- MAP kinase signaling proteins associated with cognitive decline.
- Neither protein module showed up in transcriptomic data.
Seyfried believes proteomics offers a powerful complement to RNA analysis. “We have to integrate both to get a complete picture,” he told Alzforum. “If we were completely RNA-centric, we would miss some of the strongest signals related to AD pathology.”
Commenters were impressed by the scope of the work. “This is a very comprehensive proteomic study,” Kanta Horie at Washington University in St. Louis wrote to Alzforum. Pieter Jelle Visser at Maastricht University and Betty Tijms at Amsterdam University Medical Center were intrigued by the varied biology linked to AD. “This indicates that AD affects many more processes than initially thought … We are starting to see how several processes play a role in the disease together, which is the strength of using large-scale proteomics to study disease,” they wrote (comments below).
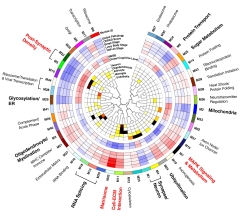
Protein Whodunit. Each protein module (outer circle) is annotated with its correlation with brain proteinopathy (next five circles; red=positive, blue=negative), cognition (next two circles), and cell-type specificity (inner circles; red to black indicates a significant association). [Courtesy of Johnson et al., Nature Neuroscience.]
More Proteins, More Networks
The authors had previously probed the AD proteome by using label-free mass spectrometry to quantify around 3,000 proteins in samples from more than 2,000 brains. That effort identified 13 protein modules, six of them linked to AD. Revved-up glial metabolism stood out as the main factor associated with the disease (May 2020 news).
To dive deeper into the proteome, joint first authors Erik Johnson, Kathleen Carter, and Eric Dammer turned to tandem-mass-tag mass spectrometry, a technique that labels proteins with chemical tags containing different heavy isotopes. This allows researchers to identify many more low-abundance proteins than do label-free methods, and also to run samples from multiple brains at once, increasing efficiency. The authors applied the technique to dorsolateral prefrontal cortex tissue from two sources, the Religious Orders Study and Memory and Aging Project (ROSMAP) and the Banner Sun Health Research Institute. Of the 516 samples, 110 came from people who had been cognitively healthy at the time of death, 206 from people with preclinical AD, and 200 from AD patients.
The tandem-mass-tag approach fished out 8,619 proteins. Three-fourths of those could be assigned to protein modules, as compared to a third of the proteins in the earlier label-free study. Modules were defined as sets of proteins that all changed together, going up or down in a coordinated fashion with disease stage. Twelve of the previously identified 13 modules, all but the smallest one, repeated in the new data. In addition, the authors found 32 new protein modules.
In some cases, previously identified modules could be refined into smaller, more specific sets. For example, proteins from the astrocyte/microglia metabolism module that dominated the first study now spread across three new modules. These were MAPK signaling (Module 7), cellular interactions with the extracellular matrix (M11), and sugar metabolism (M25). “We had such depth [of coverage] that we were able to reveal new biology,” Seyfried told Alzforum.
The findings appeared robust, replicating across paired frontal and temporal cortical samples from 113 ROSMAP brains, as well as in 151 parahippocampal gyrus tissues from the Mount Sinai Brain Bank and 40 DLPFC and anterior cingulate samples from the Emory Brain Bank.
Twelve of the 44 modules boasted particularly strong associations with plaques, tangles, and cognitive decline. For some modules, their proteins were more abundant in AD, in others, suppressed. These modules reflected processes such as synaptic function, post-synaptic density proteins (M5), sugar metabolism, myelination, RNA splicing, protein transport, and mitochondrial health. All of these had shown up in the previous study as being linked to AD. New to this study were ubiquitination, glycosylation, and the matrisome (M42). Of the 12, the four modules with the strongest associations were M5, M7, M11, and M42.
M5, M11, and M42 were altered in preclinical AD as well as in symptomatic disease, but the M7 MAPK signaling module was not. Intriguingly, M7 was also the only one of the four that remained associated with cognitive decline after adjusting for plaques and tangles. “One could postulate a model in which M42 lies ‘upstream’ in the disease cascade, whereas the M7 module may be an important effector of cognitive decline in the presence of [elevated] M42,” Johnson wrote to Alzforum.

Key Networks. Top proteins in the MAPK/metabolism (left) and the matrisome (right) modules. Yellow proteins deposit in plaques, green proteins in tangles, blue proteins in both. [Courtesy of Johnson et al., Nature Neuroscience.]
Why the RNA—Protein Disconnect ...?
To tie these findings in to previous transcriptomic work, the authors compared RNA and protein data in the same DLPFC samples from 168 of the ROSMAP brains. Surprisingly, the two overlapped only partially. Eight of the 12 AD protein modules were preserved at the RNA level, changing in the same direction in AD. Ubiquitination, glycosylation, MAPK, and the matrisome were not.
Seyfried noted that individual RNA transcripts from within these protein modules were still associated with AD. For example, most matrisome transcripts did rise with disease progression, in agreement with other work (Aug 2019 news). What was missing was the coordinated behavior of the module, where all transcripts rose or fell as a group. “There was no coherence in the biology,” Seyfried said.
One reason could be that many different cell types make and secrete matrisome proteins into the ECM. Since gene expression is regulated at the cellular level, a group of proteins with diverse cellular origins might show less concordance in their expression patterns. Once in the ECM, however, all the proteins are subject to the same forces. In particular, many of the matrisome proteins contain heparan sulfate and glycosaminoglycan-binding domains that allow them to bind Aβ fibrils, and these proteins accumulate in amyloid plaques. After becoming ensconced there, they likely last a long time, far surpassing the rapid turnover of RNA expression patterns, Seyfried noted.
Others agreed with this interpretation. “The lack of RNA upregulation may be explained by reduced clearance of the respective proteins, and/or by an accumulation in plaques and tangles,” Dietmar Thal at LU Leuven, Belgium, wrote to Alzforum (full comment below).
Likewise, the MAPK signaling proteins lack a cell-type-specific signature, and are also known to deposit in plaques and tangles. MAPK itself is a tau kinase, so this module may directly exacerbate tau pathology. This module was more strongly related to tangles than most other modules, perhaps explaining its correlation with cognition, Seyfried speculated.
… And the Gene—Protein Disconnect?
The protein modules were even less in synch with genetic polymorphisms than with transcripts. The authors looked for genetic variants that influenced the abundance of all the proteins in a module, calling these variants mod-QTLs. Perhaps unsurprisingly, they found no variants with such broad effects—with one exception. The APOE4 allele boosted levels of matrisome proteins as a set.
Again, this may have to do with protein interactions in the ECM. ApoE is in the matrisome module, as is Aβ. The E4 version could affect deposition of its fellow module proteins into plaques.
Notably, previous work has linked APOE4 to higher levels of another matrisome module protein in cerebrospinal fluid (Aug 2019 news). SMOC1, a basement membrane protein, was highly linked to AD and one of the main drivers of the module in this study, Seyfried noted.
Seyfried believes SMOC1, as well as other M7 and M42 proteins, could be biomarkers. He is extending his proteomics work to CSF and plasma to pinpoint the best candidates. Meanwhile, Johnson plans to study other neurodegenerative diseases to learn how specific the proteome changes reported here are to AD. All data from this study are freely available to other scientists.
Co-author Junmin Peng at St. Jude Children’s Research Hospital in Memphis, Tennessee, did some of the underlying proteomics analysis while at Emory. He noted the good agreement of the present findings with previous proteomics work. “This new paper demonstrates the reproducibility of large, deep proteomics datasets from multiple groups. Collectively, the results of AD proteomics will lead to innovative hypotheses to test in the future,” he wrote (full comment below).—Madolyn Bowman Rogers
References
News Citations
Further Reading
News
- Survey of Tau Partners Highlights Synaptic, Mitochondrial Roles
- Single Synapse Mass Spec Snags CD47 as Alzheimer’s Resilience Factor
- Massive Proteomics Study Connects Genes, Proteins, Disease
- Young ApoE4 Carriers Have Reversed AD Proteomic Signature
- Paper Alert: pQTLs Pin GWAS Loci to Tissue Proteins, Drug Targets
- Proteomics Dates Endosomal, Synaptic Changes to Preclinical AD
Primary Papers
- Johnson EC, Carter EK, Dammer EB, Duong DM, Gerasimov ES, Liu Y, Liu J, Betarbet R, Ping L, Yin L, Serrano GE, Beach TG, Peng J, De Jager PL, Haroutunian V, Zhang B, Gaiteri C, Bennett DA, Gearing M, Wingo TS, Wingo AP, Lah JJ, Levey AI, Seyfried NT. Large-scale deep multi-layer analysis of Alzheimer's disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat Neurosci. 2022 Feb;25(2):213-225. Epub 2022 Feb 3 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Washington University School of Medicine
This is a very comprehensive proteomic study of more than 1,000 brain tissue samples using a tandem-mass-tag mass spectrometry approach. The point of this study is that the authors leveraged brain tissue proteomics with the data from other -omics modalities, including genomics and transcriptomics, to perform a multilayer omics analysis. Ultimately, the authors revealed that a significant proportion of changes relevant to AD pathophysiology were not reflected by changes in mRNA abundance or co-expression, but could be explained by specific protein co-expression modules.
Some AD-associated protein network modules, e.g., the matrisome, were not preserved in the RNA network. Interestingly, many matrisome proteins contain heparan sulfate and glycosaminoglycan-binding domains that might mediate their interaction with amyloid plaques and potentially tau, as well. The authors also remind us that ApoE is a matrisome protein that interacts with heparan sulfate proteoglycans, and the loss of this binding interaction may be a potential mechanism for the remarkable protection afforded by the ApoE Christchurch mutation. I look forward to seeing how these large-scale proteomic analysis contribute to answering such critical questions.
One technical limitation of this study is the use of 8 M urea lysis buffer for protein extraction from brain tissue, which would recover less of, for example, membrane proteins. Also, the peptide search was conducted via “fully tryptic specificity,” which would miss proteins/peptides with such endogenous cleavages in pathophysiological conditions. Further studies using more appropriate extraction and peptide identification methods, for example de novo sequencing, will extend the findings in this report.
Maastricht University; VU University Medical Centre
Amsterdam UMC, loc. VUmc
We read this new study with a lot of pleasure. Johnson and colleagues thrust ahead and show that, for proteomic analyses, more really is more. By using tandem-mass-tag (TMT) mass spectrometry, they were able to measure five times more proteins in tissue that could be related to co-expression modules than was found with other methods. As a result, they found 44 subnetworks of proteins that co-express together in the lateral dorsal prefrontal cortex across 110 controls and 406 people with AD pathology, versus 13 that the authors had previously identified using an LFQ MS (Johnson et al., 2020). This indicates that AD affects many more processes than initially thought.
The large group of new proteins was associated with ubiquitination, glycosylation, the endoplasmatic reticulum, and the matrisome (i.e., extracellular matrix (ECM)-associated proteins). About half of the modules were also found in network analyses of RNA expression from the same sample, but not all (e.g., the MAP kinase module, which was one of the most strongly associated with pathology).
Much fundamental research focuses on RNA expression in the study of disease pathogenesis, and this finding highlights that RNA expression may not equate to protein concentration changes that are related to pathology. Each of the processes observed previously has been implied in AD, and now we are starting to see how several processes are playing a role in the disease together, which is the strength of using large-scale proteomics to study disease.
The most strongly related associations with AD pathology were postsynaptic density, MAPK signaling and metabolism, and the matrisome and cell-ECM interaction. These align with processes that we observed to be related to specific AD subtypes detected with TMT MS proteomics of the cerebrospinal fluid (Tijms et al., 2020).
In that study, we also observed a subgroup of AD patients with blood-brain barrier (BBB) disruption. The Johnson study did not find a cluster indicative of BBB dysfunction, possibly because barrier tissue was not included. It would be great if the authors would perform proteomics of the BBB in this sample, as well.
References:
Johnson EC, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, Higginbotham LA, Guajardo A, White B, Troncoso JC, Thambisetty M, Montine TJ, Lee EB, Trojanowski JQ, Beach TG, Reiman EM, Haroutunian V, Wang M, Schadt E, Zhang B, Dickson DW, Ertekin-Taner N, Golde TE, Petyuk VA, De Jager PL, Bennett DA, Wingo TS, Rangaraju S, Hajjar I, Shulman JM, Lah JJ, Levey AI, Seyfried NT. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med. 2020 May;26(5):769-780. Epub 2020 Apr 13 PubMed.
Tijms BM, Gobom J, Reus L, Jansen I, Hong S, Dobricic V, Kilpert F, Ten Kate M, Barkhof F, Tsolaki M, Verhey FR, Popp J, Martinez-Lage P, Vandenberghe R, Lleó A, Molinuevo JL, Engelborghs S, Bertram L, Lovestone S, Streffer J, Vos S, Bos I, Alzheimer’s Disease Neuroimaging Initiative (ADNI), Blennow K, Scheltens P, Teunissen CE, Zetterberg H, Visser PJ. Pathophysiological subtypes of Alzheimer's disease based on cerebrospinal fluid proteomics. Brain. 2020 Dec 1;143(12):3776-3792. PubMed.
St. Jude Children’s Research Hospital
The paper presents the most comprehensive AD proteomics analysis from brain tissues so far, with a large sample size, different brain regions, and deep proteome coverage from multiple independent cohorts. The proteomics data were generated by several proteomics groups. A number of AD proteomics studies were previously published. For example, one reported an ultra-deep analysis of brain proteome in more than 100 cases (Bai et al., 2020), while another reported a proteomic analysis of ~2,000 brains and ~400 CSF samples but with shallow proteome coverage (Johnson et al., 2020). These AD proteomic studies were recently summarized in two representative review papers (Rayaprolu et al., 2021; Bai et al., 2021).
This paper is an important continuation of the previous work. Although some of the discoveries (e.g., many matrisome proteins and the MAPK pathway) were previously described, this new paper demonstrates the reproducibility of large, deep proteomics datasets from multiple groups, significantly improving the reliability of these proteomics resources. Collectively, the results of AD proteomics have led to numerous innovative hypotheses for future investigation.
References:
Bai B, Wang X, Li Y, Chen PC, Yu K, Dey KK, Yarbro JM, Han X, Lutz BM, Rao S, Jiao Y, Sifford JM, Han J, Wang M, Tan H, Shaw TI, Cho JH, Zhou S, Wang H, Niu M, Mancieri A, Messler KA, Sun X, Wu Z, Pagala V, High AA, Bi W, Zhang H, Chi H, Haroutunian V, Zhang B, Beach TG, Yu G, Peng J. Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer's Disease Progression. Neuron. 2020 May 20;106(4):700. PubMed.
Johnson EC, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, Higginbotham LA, Guajardo A, White B, Troncoso JC, Thambisetty M, Montine TJ, Lee EB, Trojanowski JQ, Beach TG, Reiman EM, Haroutunian V, Wang M, Schadt E, Zhang B, Dickson DW, Ertekin-Taner N, Golde TE, Petyuk VA, De Jager PL, Bennett DA, Wingo TS, Rangaraju S, Hajjar I, Shulman JM, Lah JJ, Levey AI, Seyfried NT. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med. 2020 May;26(5):769-780. Epub 2020 Apr 13 PubMed.
Rayaprolu S, Higginbotham L, Bagchi P, Watson CM, Zhang T, Levey AI, Rangaraju S, Seyfried NT. Systems-based proteomics to resolve the biology of Alzheimer's disease beyond amyloid and tau. Neuropsychopharmacology. 2021 Jan;46(1):98-115. Epub 2020 Sep 8 PubMed.
Bai B, Vanderwall D, Li Y, Wang X, Poudel S, Wang H, Dey KK, Chen PC, Yang K, Peng J. Proteomic landscape of Alzheimer's Disease: novel insights into pathogenesis and biomarker discovery. Mol Neurodegener. 2021 Aug 12;16(1):55. PubMed. Correction.
Boston University School of Medicine
Levey, Seyfried, and colleagues’ proteomic studies tell us that matrisome and cell-extracellular matrix (ECM) interaction modules are significantly upregulated in early stages of Alzheimer’s disease when amyloid and tau have accumulated, and that then post-synaptic density and protein transport modules are downregulated, while MAPK signaling is upregulated, in later stages of AD when neurodegeneration may occur. Specifically, the MAPK/metabolism module highly correlates with cognitive function, whereas the matrisome does not. Interestingly, these module changes are most strongly correlated with tau level and are specific to AD but not Parkinson’s disease cases.
When they looked in the frontal cortex, roughly 50 percent of protein network modules were present in RNA networks, including synapse, protein transport and cell-ECM interaction modules, but not the matrisome. This discrepancy may be due to brain region-specific differences in RNA versus protein network consistency. Or, we do not fully understand the mechanism by which the matrisome-driven RNA network regulates protein networks, since the matrisome covers multiple submodules, including ECM, cytokines, and chemokines. Nonetheless, the matrisome protein module is highly augmented in symptomatic AD and severe AD and significantly enriched in plaque-associated proteins.
We found that the most significantly enriched pathway for distinguishing APOE 4/4 versus 3/3 is the matrisome RNA network in human mixed cortical cultures (neurons + astrocytes) derived from human iPSCs, and in brain tissue from MSBB and ROSMAP cohorts (TCW et al., 2019). We further demonstrated that the signal is derived from astrocytes when cell types are deconvoluted. Consistent with our findings, the authors found that matrisome module protein levels were enriched in ApoE ε4 protein.
The MAPK signaling and metabolism module that aligns with cognitive function is an interesting potential target for therapeutics, but we may need to further investigate the mechanism of regulation in cognitive decline using mouse and human models, and identify module proteins for use as biomarkers.
References:
Tcw J, Qian L, Pipalia NH, Chao MJ, Liang SA, Shi Y, Jain BR, Bertelsen SE, Kapoor M, Marcora E, Sikora E, Andrews EJ, Martini AC, Karch CM, Head E, Holtzman DM, Zhang B, Wang M, Maxfield FR, Poon WW, Goate AM. Cholesterol and matrisome pathways dysregulated in astrocytes and microglia. Cell. 2022 Jun 23;185(13):2213-2233.e25. PubMed. BioRxiv.
Katholieke Universiteit Leuven, Department of Imaging and Pathology, Laboratory of Neuropathology
Johnson et al. analyzed prefrontal cortex samples from 516 donors using tandem-mass-tag mass spectrometry (TMT-MS) to generate a deep TMT network of AD-related proteins. This method allows a higher resolution for the detection of proteins compared to label-free quantitation mass spectrometry. The authors found 12 modules/module families of proteins being strongly correlated with AD traits. These modules included post-synaptic density, glycosylation/endoplasmic reticulum, oligodendrocyte/myelination, RNA splicing, matrisome, cell–ECM interaction, synapse/neuron, ubiquitination, mitogen-activated protein kinase (MAPK) signaling and metabolism, mitochondria, sugar metabolism, and protein transport modules. The modules that best correlated with AD neuropathological measures (Aβ and p-tau burden) or cognitive parameters were post-synaptic density, MAPK signaling and metabolism, cell–ECM interaction, and the matrisome.
The most important finding was that two of these changed protein modules, i.e., MAPK signaling and metabolism and the matrisome, did not exhibit parallel changes in the RNA expression levels determined by transcriptomics. The authors conclude that these modules unique to the proteome could represent promising therapeutic targets or biomarkers for AD.
The matrisome covers proteins of the extracellular matrix. Their accumulation depended on APOE genotype with the ε4 allele leading to an increase. The authors identify ApoE as a member of this “matrisome” module. The very well-known accumulation of ApoE in Aβ plaques, even in very early stages of the disease, could, therefore, serve as one example that explains the lack in upregulation at the RNA level (Schmechel et al., 1993; Thal et al., 2005; Thal et al., 1997). Reduced clearance because of being trapped in plaques would be one possible explanation for such a scenario. This explanation was also supported by the Johnson et al. finding that the matrisome modules were enriched in the plaque-associated proteins.
The second proteome-unique module identified by Johnson et al. is the MAPK signaling and metabolism module. The proteins of this module are known to co-localize with plaques and tangles. Thus, the lack of RNA upregulation may also be explained by reduced clearance of the respective proteins and/or by an accumulation in plaques and/or neurofibrillary tangles together with Aβ and p-tau, respectively.
In a recent study, we compared different fractions of soluble and insoluble proteins in different stages of AD. Our results showed that the abundance of distinct proteins shifts from more-soluble into less-soluble fractions with the progression of AD (fraction-shifting proteins), supporting the pathological accumulation of these proteins results from changes in solubility and biochemical compartmentalization (Li et al., 2021).
In addition, Johnson et al. confirm earlier studies highlighting changes in synaptic proteins and protein transport modules (Johnson et al., 2020). In the context of transport processes, it is interesting to know that vesicle endocytosis, which is involved in protein transport into cells, has recently been shown to be impaired very early in the pathogenesis of AD (Li et al., 2021). Thus, it may be tempting to speculate that such transport processes could be involved in the cellular clearance of proteins that accumulate in the absence of a parallel RNA level increase.
In conclusion, Johnson et al. present results from an impressive multiomics project that shows accumulations of distinct modules of proteins that are not reflected in a parallel increase in RNA levels, suggesting that these proteins are prone either to co-aggregate with Aβ and/or p-tau, or accumulate due to impaired clearance, maybe due to decreased levels of proteins responsible for degradation, including vesicle shuttling (vesicle endocytosis) components required for this purpose.
References:
Johnson EC, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, Higginbotham LA, Guajardo A, White B, Troncoso JC, Thambisetty M, Montine TJ, Lee EB, Trojanowski JQ, Beach TG, Reiman EM, Haroutunian V, Wang M, Schadt E, Zhang B, Dickson DW, Ertekin-Taner N, Golde TE, Petyuk VA, De Jager PL, Bennett DA, Wingo TS, Rangaraju S, Hajjar I, Shulman JM, Lah JJ, Levey AI, Seyfried NT. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med. 2020 May;26(5):769-780. Epub 2020 Apr 13 PubMed.
Li X, Tsolis KC, Koper MJ, Ronisz A, Ospitalieri S, von Arnim CA, Vandenberghe R, Tousseyn T, Scheuerle A, Economou A, Carpentier S, Otto M, Thal DR. Sequence of proteome profiles in preclinical and symptomatic Alzheimer's disease. Alzheimers Dement. 2021 Jun;17(6):946-958. Epub 2021 Apr 19 PubMed.
Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9649-53. PubMed.
Thal DR, Capetillo-Zarate E, Schultz C, Rüb U, Saido TC, Yamaguchi H, Haass C, Griffin WS, Del Tredici K, Braak H, Ghebremedhin E. Apolipoprotein E co-localizes with newly formed amyloid beta-protein (Abeta) deposits lacking immunoreactivity against N-terminal epitopes of Abeta in a genotype-dependent manner. Acta Neuropathol. 2005 Nov;110(5):459-71. PubMed.
Thal DR, Glas A, Schneider W, Schober R. Differential pattern of beta-amyloid, amyloid precursor protein and apolipoprotein E expression in cortical senile plaques. Acta Neuropathol. 1997 Sep;94(3):255-65. PubMed.
Weill Cornell Medicine
This study highlights the vast difference between alterations at the protein versus transcript levels, cautioning against over-reliance on transcriptomics datasets.
The identification of a module (i.e., M7) that remains associated with cognitive decline after adjustment for neuropathology points to understudied cognitive resilience mechanisms in the presence of pathology, which could represent great opportunities for biomarkers and therapeutic strategies.
Make a Comment
To make a comment you must login or register.