Tau Filaments from the Alzheimer’s Brain Revealed at Atomic Resolution
Quick Links
Tau filaments isolated from an Alzheimer’s brain are the latest, high-profile exploit of cryo-electron microscopy. Crafting a technique that has become his claim to fame, Sjors Scheres of the MRC Laboratory of Molecular Biology in Cambridge, England, teamed up with Michel Goedert, also at MRC, to solve 3.4-Angstrom resolution structures of both straight and paired helical filaments of tau. The structures, first unveiled at the AD/PD meeting in Vienna last March, were formally published in Nature on July 5. The two filament subtypes varied at the ultrastructural level, yet shared a common C-shaped protofilament monomer made up of a series of β-sheets from the third and fourth repeat domains of tau. Two of these Cs tethered together to form each rung of the fibril. This pairing was a bit different in paired helical versus straight filaments, bestowing each fibril subtype with a distinct ultrastructural signature.

Pretty Pair.
Stacks of symmetrically paired C-shaped protofilaments generated the tau paired helical filaments found in an Alzheimer’s brain. [Courtesy of Fitzpatrick et al., Nature 2017.]
“This study represents a breakthrough in our understanding of the pathological conformation of tau,” commented Markus Zweckstetter of the Max Planck Institute for Biophysical Chemistry in Göttingen, Germany. The findings also set the stage for future cryo-EM conquests of tau fibrils from different stages of AD and in other tauopathies, he added. Researchers have been struggling to understand if different forms, or strains, of tau distinguish different tauopathies or different phases of disease progression.
First author Anthony Fitzpatrick and colleagues solved the structures by looking at the end product of tau aggregation: the filamentous tangles crowding the cortex of a 74-year-old woman who died after a 10-year history with AD. The researchers isolated tau aggregates from the sarkosyl-insoluble fraction of the tissue. As assessed initially by immuno-electron microscopy, the sample contained a mixture of paired helical filaments (PHFs) and straight filaments (SFs), the former making up about 90 percent of the total, according to Scheres. The filament mixture potently triggered aggregation of full-length tau in cultured cells. Immunoblots also revealed that both PHFs and SFs contained all six alternatively spliced isoforms of tau, including those containing all four microtubule-binding repeat domains, and those missing the second repeat (R2). Tau fibrils from the AD brain are known to contain a mixture of 3- and 4-repeat tau, while those in other tauopathies tend to contain only one or the other.

Now You C It. A C-shaped core defines the common protofilament of paired helical filaments (top) and straight filaments (bottom), though the two Cs are arranged differently. [Courtesy of Fitzpatrick et al., Nature 2017.]
Next the researchers examined the insoluble tau using cryo-EM. Fitzpatrick could see that while PHFs and SFs had different shapes, they shared a common subunit at the core (see image above). Two C-shaped structures, each formed by a molecule of tau, bound together to form individual rungs along the filament. In PHFs, the two Cs assumed a symmetrical configuration, while in SFs, these Cs were arranged slightly off-kilter (see images above). The 3.4Å resolution allowed the researchers to discern the amino acids incorporated into the fibril core. Each C-shaped structure spanned amino acids V306-F378, which contained the entire R3 and R4 repeat domains, plus an additional 10 amino acids C-terminal to the repeats.
To determine if the R3 and R4 domains were necessary and sufficient to form the core, they treated their filament samples with pronase, which is known to remove all but the cores of tau fibrils. Cryo-EM structures of treated samples still contained R3-R4, indicating that these domains formed the essential component of the fibrils. The researchers also found that an antibody specific for the R2 domain labeled filaments before, but not after, pronase treatment. This indicated that at least some filaments were composed of R2-containing 4-repeat tau, but that only the R3 and R4 domain made up the ordered core of the filament.

Double Trouble.
Tau protofilaments form when C-shaped tau cores (three shown) stack upon each other. Each core consists of eight β-sheets. The three N-terminal ones fold back on the first five to form the C. [Courtesy of Fitzpatrick et al., Nature 2017.]
The researchers presented the C-shape in striking detail. It consisted of eight β-strands, with strands 6-8 folding back on strands 1-5 to create the inside of a “two-ply” curved structure (see image above). Hydrophobic interactions and hydrogen bonds held the two plies together. The structure’s most distinctive part was an uncommon β-helical fold formed by the triangular arrangement of β sheets 4-6. This β-helix is distinct from the overall helical structure of the tau fibril. Complex β-helical folds such as these require very specific sequences to form, Goedert said. This could point to the importance of this motif, and the R4 domain that houses it, in defining the structure of AD tau fibrils, he said.
While both PHFs and SFs shared these common protofilaments, the ultrastructural differences between the two stemmed from the way they lined up with each other. In PHFs, the C-shaped subunits paired up symmetrically, tethered at the base of each C by anti-parallel interactions between the 332PGGGQ336 motif in β-sheet 3. Rock-solid hydrogen bonds stitched the Cs together.
In SFs, the two Cs joined asymmetrically and interacted weakly. Although the two subunits came close to touching via their R3 domains—at 321KCGS324 on one C, and 313VDLSK317 on the other—they were not held together via hydrogen bonds, salt bridges, or hydrophobic interactions. Instead, a region appearing as a fuzzy density on the cryo-EM appeared to span the two. The researchers speculated that this bridge could be none other than tau’s N-terminus, looping back around to stabilize the SF core (see image below). This hypothesis is based on epitopes for two tau antibodies—Alz50 and MC-1—being an amalgam of 7EFE9 in tau’s N-terminus and 313VDLSKVTSKC322 in R3, the same region where the two Cs almost touch (Carmel et al., 1996). NMR analyses of recombinant 4-repeat tau have also placed the N-terminus near the fibril core (Bibow et al., 2011). If true, then removing or mutating tau’s N-terminus should prevent the formation of SFs altogether, Scheres told Alzforum.
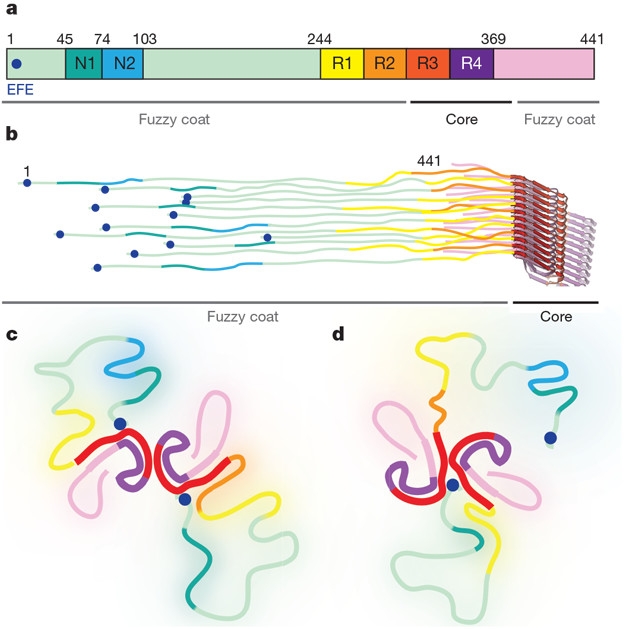
N-terminal Glue? Researchers speculated that 7EFE9 in tau’s N-terminus (blue dot, a-d), which is part of the filament’s nebulous fuzzy coat, loops back around to interact with the protofilament core (red) of PHFs (c), and SFs (d), stabilizing the latter. [Courtesy of Fitzpatrick et al., Nature 2017.]
The structure helps explain why tau fibrils in AD accommodate tau isoforms with both 3- and 4-repeats. The protofilament core only contains regions common to both types of isoform—namely the third and fourth repeat domains—so theoretically, a mixture of both isoforms could weave into the same fibril. Because tau filaments in other tauopathies contain only 3-repeat or only 4-repeat tau, Scheres and Goedert think they probably have an entirely different core. Perhaps filaments in 4-repeat tauopathies, such as progressive supranuclear palsy and corticobasal degeneration, weave the R2 domain into their core, which would exclude 3-repeat tau. The researchers are gathering postmortem brain samples from patients with other tauopathies for cryo-EM analysis.
Such differences in the fibril core could form the basis of different “strains” of tau that other researchers have observed. Marc Diamond of the University of Texas Southwestern Medical Center in Dallas has reported that tau strains, extracted from people and animals with different tauopathies and mutations, retain their distinctive physical attributes and pathogenic characteristics when passed between animals or in cell culture (May 2014 news and Nov 2016 news). Fitzpatrick’s cryo-EM structure might represent but one of these strains, commented Diamond. “Given the diversity of tau strains out there, the field collectively has a lot of work to define the many structures that no doubt exist,” he said.
The key role of the R3 domain in the AD tau fibril core dovetails with findings garnered from recombinant tau proteins, but the crucial involvement of the R4 domain was unexpected, commented Zweckstetter. That R4 contains the pivotal β-helix motif also speaks to the potential importance of the domain in fibrils from the AD brain, Scheres told Alzforum. Interestingly, the yeast prion protein Het-s also forms a β-helix, so the researchers speculated that the R4 helical motif could play a role in prion-like propagation of tau.
The researchers also proposed that the β-helix could house the binding site for tau PET tracers, which preferentially latch onto tau fibrils from the AD brain. To test this idea, they plan to generate cryo-EM structures of tau filaments bound with the AV-1451 PET ligand, Goedert said. Solving this hybrid structure, and the structures of filaments from other tauopathies, could help researchers understand how to better label and target a variety of tau strains.
David Eisenberg and Michael Sawaya of the University of California in Los Angeles also pointed out the importance of the β-helix in an accompanying commentary in Nature. “The sharp turn [β-helix] at the closed tip of the C-shape has an unexpectedly high level of structural complexity, more typical of structures that have evolved to provide some survival advantage,” they wrote. “The complexity of its pattern suggests that it could be used to develop diagnostic markers to distinguish Alzheimer’s disease from other tau-associated diseases.”
Jürgen Götz of the University of Queensland in Brisbane, Australia, agreed that the structure could help inform therapeutic strategies. “The remarkable detail in tau structure and how it differs between fibrillar and microtubule-bound tau should be useful in pursuing more targeted pharmacological strategies,” he wrote.
Goedert pointed out that the filament structures only represent the end product of tau aggregation, and tell us little about how the process began. While snipping off the protein’s “fuzzy coat” did little to alter the fibril’s core structure, this does not exclude a role for other parts of the protein in the initial formation of filaments, he said. Indeed, parts of the protein known to be hyperphosphorylated—a process that some researchers view as essential for aggregation—lie outside of the R3 and R4 domains. The role of tau fragmentation and oligomerization in kick-starting aggregation is also unclear based on the structure, he said.
The findings also question the relevance of various in vitro and cell culture models used to study tau misfolding and propagation. For example, the tau biosensor cell lines developed by Diamond express only the tau repeat domains as a templating seed, lacking the additional 10 amino acids beyond the R4 domain that were part of the fibril core in the cryo-EM structure. Other studies simply use the R3 domain to seed fibrils. “These fibrils aren’t necessarily ‘wrong,’” Scheres said. “They just may not represent what you get in the AD brain.”
Diamond called the study a wonderful contribution. “This work is inspiring us to create new types of biosensors that will encompass more of the tau protein to get more accurate representations of what is out there,” he wrote to Alzforum.—Jessica Shugart
References
Antibody Citations
News Citations
- Like Prions, Tau Strains Are True to Form
- More Evidence That Distinct Tau Strains May Cause Different Tauopathies
Paper Citations
- Carmel G, Mager EM, Binder LI, Kuret J. The structural basis of monoclonal antibody Alz50's selectivity for Alzheimer's disease pathology. J Biol Chem. 1996 Dec 20;271(51):32789-95. PubMed.
- Bibow S, Mukrasch MD, Chinnathambi S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M. The dynamic structure of filamentous tau. Angew Chem Int Ed Engl. 2011 Nov 25;50(48):11520-4. PubMed.
Further Reading
Papers
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007 May 24;447(7143):453-7. PubMed.
Primary Papers
- Fitzpatrick AW, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SH. Cryo-EM structures of tau filaments from Alzheimer's disease. Nature. 2017 Jul 13;547(7662):185-190. Epub 2017 Jul 5 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
UT Southwestern Medical Center
It is hard to overstate how important this new paper from Goedert, Scheres, and colleagues is for the Alzheimer’s disease field. From a clinical perspective, these new structures will have significant implications for how compounds are designed to detect and eliminate AD-associated tau fibrils (Kolb and Andrés, 2017). From a mechanistic perspective, this paper represents a fountainhead for a structural understanding of how different tau-associated phenotypes (or tau prion strains) arise from the structural heterogeneity of tau amyloid fibrils (Sanders et al., 2016).
Notably and surprisingly, the authors show that AD-associated tau fibrils, which include straight filaments (SFs) and paired helical filaments (PHFs), feature identical monomeric core segments and structures. In other words, differences in β-sheet alignment and amino acid identity in protease-resistant cores of the two types of filaments are not responsible for their morphological differences!…More
So what accounts for the distinctive shapes of the two types of AD-associated tau fibrils commonly seen in electron micrographs? Rather than core structure, the authors convincingly demonstrate that differences between tau amyloid filaments can be attributed to packing between cores. In other words, small tweaks in how monomers within the fibril associate with one another has drastic effects on the fibril shape. This fits amazingly well with previous work (Boluda et al., 2015; Clavaguera et al., 2013; Kaufman et al., 2016; Sanders et al., 2014) on patient-derived tau strains, which demonstrated that most AD patients likely feature a single common strain (Sanders et al., 2014), driven by a homogenous underlying amyloid structure that propagates throughout the brain to drive pathogenesis.
In other words, despite morphological differences between filaments—paired, helical, or straight—both can result from a single structure, which is defined by the identity of the core itself.
I am quite excited to see future cryo-EM-derived structures of tau fibrils obtained from postmortem tissues from patients with distinct tauopathies. Based on previous work from the Goedert, Lee/Trojanowski, and Diamond labs, it seems highly likely that distinct structures (or prion strains) will be associated with distinct diseases. Moreover, I expect to see that patients with identical syndromes will feature common structures. A structural understanding of strains associated with various diseases will provide fundamental insights into what accounts for phenotypic differences between distinct tauopathies.
With that in mind, and with my mind elsewhere in the luxurious pastures of phase separation, I’d like to pose a few questions that I look forward to the field answering:
Finally, I’d like to state that I greatly appreciate the authors’ acknowledgement that this work was only made possible by decades of support from their institution and collaborators. Solving big problems requires tremendous time, effort, resources, and perseverance. This work is evidence that such approaches to science pay enormous dividends.
References:
Boluda S, Iba M, Zhang B, Raible KM, Lee VM, Trojanowski JQ. Differential induction and spread of tau pathology in young PS19 tau transgenic mice following intracerebral injections of pathological tau from Alzheimer's disease or corticobasal degeneration brains. Acta Neuropathol. 2015 Feb;129(2):221-37. Epub 2014 Dec 24 PubMed.
Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, Probst A, Winkler DT, Reichwald J, Staufenbiel M, Ghetti B, Goedert M, Tolnay M. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci U S A. 2013 Jun 4;110(23):9535-40. PubMed.
Kaufman SK, Sanders DW, Thomas TL, Ruchinskas AJ, Vaquer-Alicea J, Sharma AM, Miller TM, Diamond MI. Tau Prion Strains Dictate Patterns of Cell Pathology, Progression Rate, and Regional Vulnerability In Vivo. Neuron. 2016 Nov 23;92(4):796-812. Epub 2016 Oct 27 PubMed.
Kolb HC, Andrés JI. Tau Positron Emission Tomography Imaging. Cold Spring Harb Perspect Biol. 2017 May 1;9(5) PubMed.
Legname G, Nguyen HO, Peretz D, Cohen FE, Dearmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci U S A. 2006 Dec 12;103(50):19105-10. PubMed.
Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, Miller TM, Grinberg LT, Seeley WW, Diamond MI. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014 Jun 18;82(6):1271-88. Epub 2014 May 22 PubMed.
Sanders DW, Kaufman SK, Holmes BB, Diamond MI. Prions and Protein Assemblies that Convey Biological Information in Health and Disease. Neuron. 2016 Feb 3;89(3):433-48. PubMed.
Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006 Aug 3;442(7102):585-9. Epub 2006 Jun 28 PubMed.
DZNE (German Center for Neurodegenerative Diseases)
This paper by Fitzpatrick et al. is a seminal accomplishment in two fields—structural biology and neurodegeneration research, both achieved at the MRC LMB in Cambridge, U.K.
For structural biology, it is an example of the recent revolution in high-resolution cryo-electron microscopy and image reconstruction. It started out ~1975 when Nigel Unwin and Richard Henderson showed that unstained cryo-preserved membrane proteins retained their native structure at atomic resolution, provided that they were imaged at low electron doses to minimize radiation damage (Unwin and Henderson, 1975). The structures could be solved by computer-based averaging over many molecules and thus revealed structures from “invisible images.” Many years of development in electron microscopy, computer and detector technology, and procedures of image processing have now opened the possibility of solving the structures of a wide range of biomolecules at high resolution, rivalling those obtained by X-ray crystallography (Scheres, 2012). …More
In neurodegeneration research, one of the key questions was the origin and structure of the “paired helical filaments” (Kidd, 1963) that appear in the brains of Alzheimer‘s patients. The major component turned out to be the microtubule-associated protein tau (Grundke-Iqbal et al., 1986). This protein had been discovered by Marc Kirschner and colleagues (Weingarten et al., 1975; Lee et al., 1988), who noted two key properties, stabilization of axonal microtubules and an unusually hydrophilic composition, making it the prototype of a natively unfolded and highly soluble protein. It is therefore still a mystery why this protein aggregates into semi-ordered filaments, yet the distribution of these filaments in the brain serves as a reliable marker of the progression of AD (the “Braak stages” proposed by Heiko and Eva Braak, 1991). In several important papers around 1988 onwards, the group at LMB Cambridge (Aaron Klug, Claude Wischik, Michel Goedert, Tony Crowther, and colleagues) described key structural and biochemical properties of tau and tau filaments, including the six isoforms of human tau, the core of PHFs and their “fuzzy coat,” and low-resolution reconstructions from electron micrographs of negatively stained tau fibers from Alzheimer brain (PHF and straight filaments). Three decades later, the new publication confirms the basic conclusion and extends them to near-atomic resolution of the core of tau fibers.
Our lab has been active in tau research since ~1988, much of it on structural, biochemical, and cellular aspects of tau. The new structure of Fitzpatrick et al. now offers a chance to revisit earlier views, as discussed below:
Core of PHFs and amyloidogenic motifs: The tau sequence contains two hexapeptide motifs with high β-propensity, 306-VQIVYK-311 at the beginning of R3 (termed PHF6) and 275-VQIINK-280 (=PHF*) at the beginning of R2 (von Bergen et al., 2000). Their extended conformation is already visible by NMR in soluble tau (Mukrasch et al., 2009), and the corresponding peptides can be crystallized to reveal the amyloid-like packing into cross-β-sheets (Sawaya et al., 2007). Both motifs strongly promote aggregation of tau, yet the new structure reveals only the motif VQIVYK in R3. Thus it appears that there is another β-structured part in the filament comprising R2 which may be too heterogeneous to show up in the map. Note that in the filaments from this patient, R2 is present in only 50 percent of the tau molecules because of alternative splicing of exon 10 (encoding R2). However, even filaments consisting entirely of 4-repeat tau show this heterogeneity, in contrast to R3, as judged by site-directed EPR (Margittai and Langen, 2006). The missing β-structured part of R2 might become visible in future studies of tau filaments from 4R-tauopathies. A surprising feature is the pronounced appearance of repeats R4 and R5. R4 also harbors an amyloigenic hexapeptide motif, 337-VEVKSE-342 (=PHF6**), with an extended conformation already noticeable in soluble tau. The sequence of R5,also termed R' (Gustke et al., 1994) shows a lower sequence homology than the other repeats, but its initial hexapeptide KKIET also shows up as β-structure in the map. Frequently used recombinant constructs comprising the repeat domains such as K19 (=R1+R3+R4) or K18 (=R1+R2+R3+R4) end with the motif 369-KKIE-372 of repeat R5, which may be one reason why they can be assembled into filaments resembling PHFs.
Tau mutations: Most of the mutations known to cause tauopathies in humans occur around the repeat domain, in particular in or near the hexapeptide motifs. One likely mode of action is to enhance the tendency for tau aggregation. Examples are N279K, ΔK280 (in R2), P301L, P301S (just upstream of R3), V337M (in R4), K369I (in R5). Alternatively, these mutations could affect the binding of tau to microtubules or other tau effectors, since the hexapeptide motifs are also involved in those interactions (Kadavath et al., 2015).
Protofilaments: The term "paired helical filament" was coined by Kidd to describe the appearance of stained filaments from AD brain tissue. However, a fraction of filaments did not have the twisted appearance and were therefore called "straight filaments" (Crowther, 1991). Since the packing of tau molecules was not known there has been a debate in the literature on whether the straight filaments (SF) consisted of one or two protofibrils (Pollanen et al., 1997; Wegmann et al., 2010), especially since it was not possible to observe protofilaments fraying apart (unlike, say, the case of microtubules, where fraying protofilaments are readily visible). The new structure puts the issue to rest by clearly revealing two protofilaments, consistent with Crowther's 1991 image reconstruction.
Mass distribution: Because tau is highly soluble it does not self-assemble readily, except in special conditions (Wille et al., 1992) or with cofactors distinct from the cellular milieu (Goedert et al., 1996). The fibers assembled in vitro show some heterogeneity, which caused a debate whether they had the same packing of molecules as PHFs from AD brain tissue. We therefore determined the molecular mass per length of AD-PHFs and filaments reassembled in vitro from recombinant tau with different domain compositions (von Bergen et al., 2006). This showed that all filaments contained ~four molecules per nm length, independent of the size of the tau subunits. This agrees well with the structure of Fitzpatrick et al., which contains two protofilaments, with 0.47nm spacing between molecules in each.
Oxidation of tau: Tau contains either one cysteine (C322 in 3-repeat tau) or two (C291+C322 in 4-repeat tau). Cells have a reducing environment, but aging cells are compromised such that oxidation can occur. When studying the effects of oxidation we noticed that 4-repeat tau assembles poorly, whereas 3-repeat tau assembles much more readily than controls where the cysteines had been substituted by Ala. The explanation was that oxidized 4-repeat tau assumes a folded conformation via an intrachain disulfide crosslink between C291 and C322 which is not well integrated into a fiber, whereas oxidized 3-repeat tau can form interchain disulfide bridges to generate dimers and then polymers (Schweers et al., 1995). This interpretation is consistent with the new structure: Although it shows only C322 (because R2 is not resolved), its position precludes an interaction with C291 in the assembled filament. By contrast, C322 is able to make a disulfide bond with another molecule in the axial direction, thus promoting the templated assembly of the filament. The existence of such a disulfide bond has been verified by solid state NMR (Daebel et al., 2012). By implication, the structure also illustrates how blocking of SH goups (e.g., by the aggregation inhibitor methylene blue) can antagonize the formation of PHFs (Akoury et al., 2013).
Phosphorylation of tau: In neurons, aggregation of tau is usually accompanied by phosphorylation, but does not depend on it. Tau contains multiple phosphorylation sites, most of them outside the repeat domain. The predominant sites within the repeats are the serines in the KXGS motifs (X=I or C) in R1-R4 (phosphorylation mainly of S262, S356 in the KIGS motifs, whereas S293, S324 in the KCGS motifs are minor sites). They can be targeted by kinases MARK, SADK, PKA, and others, and all sites are recognized by the antibody 12E8 generated by Peter Seubert at Elan. This type of phosphorylation, especially at S262, reduces tau-microtubule interactions as well as tau aggregation (Schneider et al., 1999). The new map shows only residues S324 (in R3) and S356 (in R4) in an unphosphorylated form. S324 faces outside of the C-shaped structure, consistent with its minor role in inhibiting PHF assembly. S356 lies on the inside of the C-shape and appears to be tightly squeezed between charged residues K354 and D358, suggesting that phosphorylation could disrupt the conformation and thus PHF assembly. The environment of S262 remains to be elucidated.
Non-tau components of PHFs: Recombinant tau protein is soluble at concentrations >200 µM, even in a highly phosphorylated state (Tepper et al., 2014). On the other hand, neuronal tau is in the low µM range, ~10-fold lower than tubulin (Drubin et al., 1984). Therefore, tau by itself would not assemble in cells nor in vitro in physiological buffer conditions unless it interacts with a cellular partner that compensates its excess positive charges, especially in the repeat domain that harbors the elements for β-structure. In vitro, filament aggregation can be achieved by polyanions such as heparin (Goedert et al., 1996), RNA, or acidic peptides (Friedhoff et al., 1998). The acidic C-terminal tail of tubulin would qualify as a partner for tau aggregation, except that microtubules act as chaperones of tau. Erickson and Voter likened the tubulin-tau interaction to the complex coacervation of polyelectrolytes, where assembly of (acidic) tubulin is induced by (basic) polycations, e.g. tau (Erickson and Voter, 1976). Reversing the argument, the assembly of (basic) tau can be induced by (acidic) polyanions. In vitro, heparin serves as an inducer, but in the cytoplasm there must be alternatives. Examples are the abundant RNAs (rRNA, mRNA, tRNA). Indeed, stress granules containing RNA and proteins including tau can be viewed as one type of complex coacervate (Vanderweyde et al., 2016). The question is: Where are the extra components in the PHF map? The ordered elements reflect only a part of tau itself. It is possible that other components are hidden in the disordered background, or that they were removed by proteolysis during aging or preparation. In any case, the secret to understanding pathological tau aggregation might well lie in the polyanionic binding partner(s), rather than just in tau.
In 1903, when Alois Alzheimer moved to the University Clinic in Munich headed by Emil Kraepelin, he declined to become a regular professor with teaching duties, but instead asked for a room full of microscopes to pursue the research for which he became famous. One hundred and fourteen years and a huge progress in microscopy methods later, the new structure of the protein that forms neurofibrillary tangles is a fitting follow-up that will propel future directions of research in neurodegenerative diseases.
References:
Unwin PN, Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975 May 25;94(3):425-40. PubMed.
Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012 Jan 13;415(2):406-18. Epub 2011 Nov 12 PubMed.
Grundke-Iqbal I, Iqbal K, Quinlan M, Tung YC, Zaidi MS, Wisniewski HM. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem. 1986 May 5;261(13):6084-9. PubMed.
Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci U S A. 1975 May;72(5):1858-62. PubMed.
Lee G, Cowan N, Kirschner M. The primary structure and heterogeneity of tau protein from mouse brain. Science. 1988 Jan 15;239(4837):285-8. PubMed.
Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239-59. PubMed.
von Bergen M, Friedhoff P, Biernat J, Heberle J, Mandelkow EM, Mandelkow E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc Natl Acad Sci U S A. 2000 May 9;97(10):5129-34. PubMed.
Mukrasch MD, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009 Feb 17;7(2):e34. PubMed.
Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007 May 24;447(7143):453-7. PubMed.
Margittai M, Langen R. Side chain-dependent stacking modulates tau filament structure. J Biol Chem. 2006 Dec 8;281(49):37820-7. PubMed.
Gustke N, Trinczek B, Biernat J, Mandelkow EM, Mandelkow E. Domains of tau protein and interactions with microtubules. Biochemistry. 1994 Aug 16;33(32):9511-22. PubMed.
Kadavath H, Jaremko M, Jaremko Ł, Biernat J, Mandelkow E, Zweckstetter M. Folding of the Tau Protein on Microtubules. Angew Chem Int Ed Engl. 2015 Aug 24;54(35):10347-51. Epub 2015 Jun 19 PubMed.
Crowther RA. Straight and paired helical filaments in Alzheimer disease have a common structural unit. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2288-92. PubMed.
Pollanen MS, Markiewicz P, Goh MC. Paired helical filaments are twisted ribbons composed of two parallel and aligned components: image reconstruction and modeling of filament structure using atomic force microscopy. J Neuropathol Exp Neurol. 1997 Jan;56(1):79-85. PubMed.
Wegmann S, Jung YJ, Chinnathambi S, Mandelkow EM, Mandelkow E, Muller DJ. Human Tau isoforms assemble into ribbon-like fibrils that display polymorphic structure and stability. J Biol Chem. 2010 Aug 27;285(35):27302-13. PubMed.
Wille H, Drewes G, Biernat J, Mandelkow EM, Mandelkow E. Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J Cell Biol. 1992 Aug;118(3):573-84. PubMed.
Goedert M, Jakes R, Spillantini MG, Hasegawa M, Smith MJ, Crowther RA. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature. 1996 Oct 10;383(6600):550-3. PubMed.
von Bergen M, Barghorn S, Müller SA, Pickhardt M, Biernat J, Mandelkow EM, Davies P, Aebi U, Mandelkow E. The core of tau-paired helical filaments studied by scanning transmission electron microscopy and limited proteolysis. Biochemistry. 2006 May 23;45(20):6446-57. PubMed.
Schweers O, Mandelkow EM, Biernat J, Mandelkow E. Oxidation of cysteine-322 in the repeat domain of microtubule-associated protein tau controls the in vitro assembly of paired helical filaments. Proc Natl Acad Sci U S A. 1995 Aug 29;92(18):8463-7. PubMed.
Daebel V, Chinnathambi S, Biernat J, Schwalbe M, Habenstein B, Loquet A, Akoury E, Tepper K, Müller H, Baldus M, Griesinger C, Zweckstetter M, Mandelkow E, Vijayan V, Lange A. β-Sheet core of tau paired helical filaments revealed by solid-state NMR. J Am Chem Soc. 2012 Aug 29;134(34):13982-9. PubMed.
Akoury E, Pickhardt M, Gajda M, Biernat J, Mandelkow E, Zweckstetter M. Mechanistic basis of phenothiazine-driven inhibition of tau aggregation. Angew Chem Int Ed Engl. 2013 Mar 18;52(12):3511-5. PubMed.
Tepper K, Biernat J, Kumar S, Wegmann S, Timm T, Hübschmann S, Redecke L, Mandelkow EM, Müller DJ, Mandelkow E. Oligomer formation of tau protein hyperphosphorylated in cells. J Biol Chem. 2014 Dec 5;289(49):34389-407. Epub 2014 Oct 22 PubMed.
Drubin DG, Caput D, Kirschner MW. Studies on the expression of the microtubule-associated protein, tau, during mouse brain development, with newly isolated complementary DNA probes. J Cell Biol. 1984 Mar;98(3):1090-7. PubMed.
Goedert M, Jakes R, Spillantini MG, Hasegawa M, Smith MJ, Crowther RA. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature. 1996 Oct 10;383(6600):550-3. PubMed.
Friedhoff P, Schneider A, Mandelkow EM, Mandelkow E. Rapid assembly of Alzheimer-like paired helical filaments from microtubule-associated protein tau monitored by fluorescence in solution. Biochemistry. 1998 Jul 14;37(28):10223-30. PubMed.
Erickson HP, Voter WA. Polycation-induced assembly of purified tubulin. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2813-7. PubMed.
Vanderweyde T, Apicco DJ, Youmans-Kidder K, Ash PE, Cook C, Lummertz da Rocha E, Jansen-West K, Frame AA, Citro A, Leszyk JD, Ivanov P, Abisambra JF, Steffen M, Li H, Petrucelli L, Wolozin B. Interaction of tau with the RNA-Binding Protein TIA1 Regulates tau Pathophysiology and Toxicity. Cell Rep. 2016 May 17;15(7):1455-1466. Epub 2016 May 6 PubMed.
Max Planck Institute for Biophysical Chemistry
This study represents a breakthrough in our understanding of the pathological conformation of the Alzheimer-related protein tau. In a unique manner it combines preparations of insoluble protein deposits from a patient with Alzheimer’s disease, which were first discovered more than 30 years ago, with technical advances in structural biology, in particular in single-particle cryo-EM. The study provides high-resolution insights into the conformation of amyloid fibrils of tau. In agreement with previous studies, it shows that the pseudo-repeat R3, which is the most hydrophobic region of the tau sequence, is part of the core of tau fibrils.
Rather unexpectedly, however, the β-structure-containing region, which is well-defined in the electron density maps, includes the complete R4 region and even extends to F378 downstream of R4. In liquid-state NMR measurements of fibrils of 2N4R tau prepared in vitro (Bibow et al., 2011; Sillen et al., 2005), residues starting at ~400 become observable, suggesting that the structured region of tau amyloid fibrils might even extend beyond F378. In addition, residues in R2 and even R1 might contribute to the fibril core, although the definition of electron density in these regions was not sufficient to make that determination—potentially due to the presence of multiple isoforms, which either have or don’t have R2, in the patient-derived material.…More
Another interesting question is how much variation in the local β-structure can occur in different preparations of tau fibrils purified from patients, given the observation that a significant amount of heterogeneity was observed in heparin-induced tau fibrils of K18/K19 by solid-state NMR spectroscopy (Andronesi et al., 2008; Daebel et al., 2012; Xiang et al., 2017). The observation of additional electron density, which cannot unambiguously be assigned to specific amino acids in tau, further stresses the need to integrate cryo-EM studies of patient-derived material with NMR studies of in vitro aggregated amyloid fibrils. This is, for example, highlighted by the observation of additional electron density, which might originate from the very N-terminus of tau consistent with an interaction of this region with the amyloid core suggested by liquid-state NMR studies (Bibow et al., 2011). In addition, in vitro-prepared amyloid fibrils of tau, will enable the study of the influence of site-directed mutations and post-translational modifications on the structure of aggregated states of the protein. The study forms the basis for characterization of the structure of insoluble deposits from other patients with AD, or those in different stages of AD potentially including presymptomatic AD, as well as from other forms of tauopathy.
References:
Bibow S, Mukrasch MD, Chinnathambi S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M. The dynamic structure of filamentous tau. Angew Chem Int Ed Engl. 2011 Nov 25;50(48):11520-4. PubMed.
Sillen A, Leroy A, Wieruszeski JM, Loyens A, Beauvillain JC, Buée L, Landrieu I, Lippens G. Regions of tau implicated in the paired helical fragment core as defined by NMR. Chembiochem. 2005 Oct;6(10):1849-56. PubMed.
Andronesi OC, von Bergen M, Biernat J, Seidel K, Griesinger C, Mandelkow E, Baldus M. Characterization of Alzheimer's-like paired helical filaments from the core domain of tau protein using solid-state NMR spectroscopy. J Am Chem Soc. 2008 May 7;130(18):5922-8. PubMed.
Daebel V, Chinnathambi S, Biernat J, Schwalbe M, Habenstein B, Loquet A, Akoury E, Tepper K, Müller H, Baldus M, Griesinger C, Zweckstetter M, Mandelkow E, Vijayan V, Lange A. β-Sheet core of tau paired helical filaments revealed by solid-state NMR. J Am Chem Soc. 2012 Aug 29;134(34):13982-9. PubMed.
Xiang S, Kulminskaya N, Habenstein B, Biernat J, Tepper K, Paulat M, Griesinger C, Becker S, Lange A, Mandelkow E, Linser R. A Two-Component Adhesive: Tau Fibrils Arise from a Combination of a Well-Defined Motif and Conformationally Flexible Interactions. J Am Chem Soc. 2017 Feb 22;139(7):2639-2646. Epub 2017 Feb 8 PubMed.
Make a Comment
To make a comment you must login or register.