Plasma Proteomics Study Hints at New Player in Alzheimer’s
Quick Links
Sifting plasma for hundreds of proteins that foster cell-to-cell communication, scientists led by Tony Wyss-Coray at Stanford University School of Medicine, California, appear to have fished out a heretofore unknown signaling pathway linked to Alzheimer’s disease. As the scientists report in the April 26 Molecular Neurodegeneration, AD patients appear to have less growth differentiation factor signaling in the brain, a result that finds support in genomic and transcriptomic analysis. The authors believe their multi-tiered approach may uncover other pathways associated with Alzheimer’s disease—ones that might be targeted therapeutically.
“This is a massive amount of work,” said Henrik Zetterberg, University of Gothenburg, Mölndal, Sweden. This type of analysis—combining proteomics with bioinformatics, tissue studies, and culture studies—might eventually reveal strong plasma biomarkers, he said.
In 2007, Wyss-Coray and colleagues described a panel of 18 plasma proteins that could distinguish AD from control samples and predict conversion from mild cognitive impairment to AD (Oct 2007 news). Other groups have been unable to replicate those findings, and the search is still on for a blood test for Alzheimer’s almost a decade later (Feb 2016 news; Jun 2013 webinar).
This time around, Wyss-Coray aimed to find plasma biomarkers that would serve another purpose—to highlight novel pathways involved in AD. They focused on circulating signaling proteins, a subgroup they chose based on the idea that floundering neurons send out distress calls that cross a compromised blood-brain barrier. “If we can eavesdrop on that communication, then we might learn about the stress state of the brain,” said first author Philipp Jaeger, now at the University of California, San Diego.
Their goal, Jaeger explained, was to find individual proteins that differed between AD and healthy aging, and then collapse those lone hits onto common signaling pathways. The idea was that while individual patients might be affected at different points in a given signaling cascade, collecting hits across patient populations and assigning them to pathways might reveal common ones that come under fire in disease.
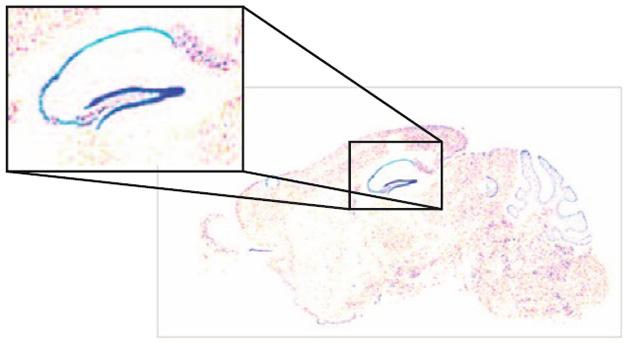
Focused Expression. GDF3 (blue) is highly expressed in the dentate gyrus of the mouse. [Courtesy of Jaeger et al., 2016, http://creativecommons.org/licenses/by/4.0.]
To start out, Jaeger and colleagues examined blood samples from 47 people with sporadic AD and 52 age-matched controls collected at the University of California, San Francisco, and the Mayo Clinics in Rochester, Minnesota, and Jacksonville, Florida. Antibody-based microarray analysis revealed which of about 600 signaling proteins varied with disease.
The authors analyzed the results three ways. First, they compared concentrations between AD and control subjects and found 77 proteins in three broad functional categories: apoptosis, complement activation, and TGFβ/GDF/BMP signaling. The latter controls growth and differentiation in embryonic and adult cells. Second, to figure out which of the 600 proteins best correlated with disease progression, the researchers compared levels with donors’ MMSE scores. Once again, this analysis pulled up factors related to TGFβ/GDF/BMP signaling and apoptosis. Proteins related to complement activation did appear to track with MMSE, but the correlation was weaker. Lastly, the authors did a co-expression analysis, where they looked at how protein pairs co-vary in disease. Once again, three main clusters appeared to be altered in parallel: complement, growth regulation, and apoptosis.
To check whether this experimental approach pulled out proteins linked to AD brain pathology, Jaeger looked to the published literature. Researchers led by Eric Schadt at the Icahn School of Medicine at Mount Sinai in New York and Valur Emilsson at the Icelandic Heart Association, in Kopavogur had recently correlated mRNA transcripts in brain tissue from AD patients with Braak staging and the degree of brain atrophy (see May 2013 webinar on Zhang et al., 2013). Almost a third of the proteins Jaeger and colleagues correlated with MMSE score also correlated with Braak staging in Schadt’s study, and another third tracked with atrophy. This suggested that a focus on secreted signaling molecules could enrich for those related to AD brain pathology.
Since apoptosis and the complement pathway are well-studied in AD, the researchers decided to narrow their focus to the TGFβ superfamily to find novel markers. TGFβ signaling has been studied in AD, but the GDF subgroup has not been associated with the disease. The researchers pored over metadata from 10 genome-wide association studies (GWAS) and AD brain transcriptome data from 181 AD patients and 125 controls to see whether SNPs or mRNA changes in the TGFβ pathway associate with AD. They found a higher degree of SNPs and altered mRNA expression than would be expected by chance.
Growth differentiation factor 3 (GDF3) came up repeatedly. This protein is highly expressed in the human brain and concentrates in the dentate gyrus in mice (see image above), but its effects on neurons are unknown. Since the authors had found reduced GDF3 in the patient plasma, they checked its levels in postmortem brain tissue. They found less in the cortices of AD patients than controls. Given its high expression in the mouse dentate gyrus, they suspected it could have an impact on neurogenesis. Sure enough, primary neural progenitor cells cultured from mice differentiated into neurons and astrocytes more efficiently when they were exposed to GDF3. The factor also promoted neuronal differentiation in pluripotent human embryonal carcinoma cells. Taken together, the results suggest that loss of GDF signaling may play a role in AD by limiting neurogenesis.
“The study represents a beautiful hierarchical strategy that is internally highly consistent,” said Terrence Town, University of Southern California, Los Angeles, who was surprised that the TGFβ superfamily came up as such a strong hit. “Everything points to the TGFβ superfamily as being at the epicenter of these proteomic changes, and supports the idea that TGFβ signaling is dysfunctional in AD,” Town added. Town’s previous work suggests that unusually high TGFβ signaling in the AD brain subdues microglial activity that would otherwise help clear AD pathology (Jun 2008 news). That a reduced amount of GDF3 signaling may play into AD speaks to the cell-specific effects of this family of proteins.
For his part, Zetterberg said he would have liked to see more accurate measures to validate the protein changes suggested by the microarray, before the researchers did further analyses. Methods such as ELISA could give better quantitative reads for the top hits, he said. He also would like to see these results replicated in another patient sample. Future studies might test whether the same proteins change in people with early disease, perhaps those diagnosed with prodromal AD, he said.—Gwyneth Dickey Zakaib
References
News Citations
- A Blood Test for AD?
- Replication a Challenge in Quest for Alzheimer’s Blood Test
- Macrophages Storm Blood-brain Barrier, Clear Plaques—or Do They?
Webinar Citations
- Webinar: O Blood-Based Biomarker, Where Art Thou?
- Can Network Analysis Identify Pathological Pathways in Alzheimer’s
Paper Citations
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, MacDonald ME, Lamb JR, Bennett DA, Molony C, Stone DJ, Gudnason V, Myers AJ, Schadt EE, Neumann H, Zhu J, Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013 Apr 25;153(3):707-20. PubMed.
Further Reading
Papers
- Chiam JT, Dobson RJ, Kiddle SJ, Sattlecker M. Are blood-based protein biomarkers for Alzheimer's disease also involved in other brain disorders? A systematic review. J Alzheimers Dis. 2015;43(1):303-14. PubMed.
- Kim DH, Lee D, Chang EH, Kim JH, Hwang JW, Kim JY, Kyung JW, Kim SH, Oh JS, Shim SM, Na DL, Oh W, Chang JW. GDF-15 secreted from human umbilical cord blood mesenchymal stem cells delivered through the cerebrospinal fluid promotes hippocampal neurogenesis and synaptic activity in an Alzheimer's disease model. Stem Cells Dev. 2015 Oct 15;24(20):2378-90. Epub 2015 Aug 19 PubMed.
Primary Papers
- Jaeger PA, Lucin KM, Britschgi M, Vardarajan B, Huang RP, Kirby ED, Abbey R, Boeve BF, Boxer AL, Farrer LA, Finch N, Graff-Radford NR, Head E, Hofree M, Huang R, Johns H, Karydas A, Knopman DS, Loboda A, Masliah E, Narasimhan R, Petersen RC, Podtelezhnikov A, Pradhan S, Rademakers R, Sun CH, Younkin SG, Miller BL, Ideker T, Wyss-Coray T. Network-driven plasma proteomics expose molecular changes in the Alzheimer's brain. Mol Neurodegener. 2016 Apr 26;11:31. PubMed. Correction.
Annotate
To make an annotation you must Login or Register.

Comments
National Institute on Aging
Traditional biomarker discovery studies in AD have relied upon a case–versus-control design wherein the binary discrimination of presence/absence of disease based on single analytes is the primary outcome. There are several limitations to this approach, including ignoring the significant levels of AD pathology in non-demented controls and a long preclinical prodrome of AD where pathology is accumulating in the brain prior to symptom onset. These studies also ignore the considerable heterogeneity in disease progression and symptoms in AD patients. For these reasons, single analyte markers arising from these case versus control studies have been notoriously difficult to replicate.
The novel aspect of the present study is the use of a network approach, wherein both differential expression of individual proteins as well as the co-expression of multiple proteins within networks representing biological pathways relevant to AD pathogenesis are considered important. Secondly, the authors have used a systems-level approach, integrating data from multiple publicly available genome-wide association studies as well as gene expression datasets from brain to demonstrate a biological validation of their biomarker signals. Thirdly, rather than relying entirely on the binary discrimination between cases and controls, the authors used an endophenotype, i.e., a measure of global cognition represented by the MMSE score as an outcome variable to test associations with their biomarkers of interest.…More
The study design is novel and cleverly leverages large datasets in the public domain to establish biologically plausible protein markers of AD arising from their index antibody array experiments. These pathways contain several of the “usual suspects” reported previously in other AD biomarker studies such as Complement C3, Haptoglobin, and Complement Factor H—a fact that provides a measure of confidence in the robustness of the results. A novel protein marker, GDF, also emerges from these analyses and merits independent validation in larger samples from different cohorts.
In order to reach clinical utility, such biomarkers require extensive independent validation. The immediate implications of studies such as this are the expansion of our knowledge of novel disease mechanisms that may potentially be targets for disease modification in AD.
Commonwealth Scientific and Industrial Research Organisation
The recent report by Jaeger and colleagues represents an exciting addition to current knowledge on plasma markers for AD. They present a comprehensive multi-level analysis pipeline to identify three biological pathways likely associated with AD: TGFβ/GDF/BMP signaling, apoptosis, and the complement system. By comparing and contrasting their results to those discovered in an alternative neurodegenerative disease (semantic-variant primary progressive aphasia, a sub-type of frontotemporal lobe dementia) they provide an essential validation of the findings; however, extension of this to other neurodegenerative diseases would be of equal importance in assessing validity. With early, prognostic identification of AD sufferers representing the “Holy Grail” for plasma biomarkers, further longitudinal evaluation will be required to gain a true indication of the potential clinical efficacy represented by these findings.…More
Make a Comment
To make a comment you must login or register.