In Mice, Gene Editing Neutralizes Mutant APP, Keeps Plaques Away
Quick Links
Could CRISPR gene-editing technology be used to cure autosomal-dominant Alzheimer’s disease? The question preoccupies both families and scientists concerned with ADAD. Researchers led by Nancy Ip at Hong Kong University of Science and Technology reported that it can, at least in mice. In the July 26 Nature Biomedical Engineering, they described an adeno-associated virus carrying gene-editing tools that disrupt mutated amyloid precursor protein gene, while leaving wild-type alleles intact. A single injection of the AAV into the brains of mouse models of APP amyloidosis cut plaque accumulation over six months by 80 percent. Gliosis and neurite dystrophy also slowed. Intravenous infusion of a different AAV that crosses the blood-brain barrier achieved the same effect, and even tempered cognitive deficits.
- AAV-based CRISPR targeted mutant APP, limiting plaques in mice. Viruses that infiltrate only the CNS—and only neurons there—may facilitate localized gene editing in the brain.AAV9 carrying CRISPR cuts mutant APP, spares wild-type, in mice.
- Halts plaques, gliosis, neurite dystrophy, cognitive decline.
- Newly engineered AAVs better target the brain.
A team led by Viviana Gradinaru, Caltech, Pasadena, California, in collaboration with Ip, reported in the December 9 Nature Neuroscience their creation of new AAVs that cross the BBB and infect the brain, and only neurons there, while sparing peripheral organs. Achieving this specificity from peripheral delivery creates new research and therapeutic opportunities unattainable with currently available AAVs.
“This is an exciting example of what gene therapy for an intractable neurological condition could be,” Olivier Danos of Maryland-based REGENXBIO wrote to Alzforum. Subhojit Roy, University of California, San Diego, agreed. “Though applicable to a small fraction of Alzheimer’s patients carrying the familial Swedish mutation, [Ip’s study] opens the door for similar ‘personalized’ CRISPR-based approaches for other Alzheimer’s mutations,” he wrote to Alzforum. “It is difficult to imagine a future where gene-based therapies will not be translated to more common neurodegenerative diseases,” Roy added. As for Gradinaru’s contribution, Martin Ingelsson, University of Toronto, wrote, “This elegant ... work represents an important step forward to enable future AAV-based systemic therapies for various brain disorders” (full comments below).
With CRISPR gene editing, researchers can change specific genetic sequences with great precision. For example, scientists led by Ingelsson previously nicked the APP gene near the Swedish KM670/671NL double mutation to induce a frameshift and remove the nearby BACE cleavage site. This diminished Aβ production in cultured cells from ADAD patients; alas, it edited so few cells in the brains of transgenic mice—below 2 percent of neurons—that the scientists did not assess any effect on AD pathology (May 2018 news). Similarly, South Korean researchers used CRISPR nanocomplexes to suppress BACE1 expression, which reduced amyloid plaques and improved learning and memory (Mar 2019 news).
Now, co-first authors Yangyang Duan and Tao Ye from Ip’s lab achieved the same effect by packaging CRISPR machinery into AAV viruses. Similar to Ingelsson, the Hong Kong group created a guide RNA targeting 27 nucleotides surrounding the Swedish APP mutation. In CRISPR-transfected HEK cells, the guide RNA drove Cas9 to cut only mutant APP, leaving the wild-type allele intact (see image below).
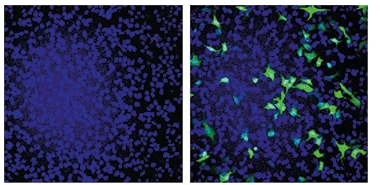
Cut It Out. In HEK cells, a guide RNA directed Cas9 to cut a snippet of APP containing the Swedish mutation from within a green fluorescent protein gene, restoring production of intact GFP (right). A wild-type APP fragment/GFP chimera stayed intact (left). [Courtesy of Duan et al., Nature Biomedical Engineering, 2021.]
With this guide RNA in hand, the scientists packaged it and Cas9 mRNA into AAV9, a serotype commonly used to deliver gene therapies to the CNS (reviewed by Saraiva et al., 2016). They injected the virus into one side of the hippocampi of 3-month-old 5xFAD mice, which carry five familial AD mutations, including the Swedish APP mutation. Four weeks later, 27 percent of hippocampal cells from the treated side were devoid of the Swedish mutation, indicating it had been edited out. Danos noted that this is a surprisingly high level of gene editing.
Did this matter, though? At 3 months of age, 5xFAD mice begin accumulating plaques; and plaque deposition, gliosis, and neuronal dysfunction are widespread by 6 months. In treated 5xFAD mice, the scientists measured 65 to 80 percent less soluble and insoluble Aβ40 and Aβ42, and a 78 percent lower plaque load in the ipsilateral hippocampus compared to the un-injected control side (see image below). This reduction held by 9 months of age.

CRISPR Prevents Plaques. Three months after AAV9-CRISPR injection, 6-month-old 5xFAD mice had fewer Aβ deposits (green) in the treated (bottom) than untreated (top) side of their hippocampus. Nuclei are blue; hemagglutinin-tagged Cas9, red. [Courtesy of Duan et al., Nature Biomedical Engineering, 2021.]
CRISPRed mice also had 35 percent fewer Iba1-positive microglia at 6 months, while the number of their astrocytes stayed unchanged. Long-term potentiation (LTP), a sign of synaptic plasticity, came in near wild-type levels, suggesting less neurotoxicity than in untreated controls. In keeping with this, tissue sections from treated mice had 44 percent more PSD-95-positive puncta, 20 percent more NeuN-positive neurons, and half as many dystrophic neurites as did tissue from untreated animals.
To see how gene editing might affect more advanced pathology, the scientists injected the AAV-CRISPR into the hippocampi of 9-month-old APP/PS1 mice, which carry the Swedish APP plus one PSEN1 mutation and have extensive plaque burden and gliosis by 9 months of age. At 15 months, treated mice contained half as many hippocampal plaques, half as many activated microglia, and 18 percent fewer GFAP-positive astrocytes than controls.
Could systemic treatment achieve a brain-wide reduction of pathology? The scientists packaged the same guide RNA and Cas9 mRNA into a BBB-penetrating variant of AAV9, called AAV-PHP.eB (Chan et al., 2017; Nov 2019 conference news). They intravenously injected 3-month-old 5xFAD mice, which achieved similar editing efficiency in the brain as did the intrahippocampal injection. Cas9 was expressed throughout the brain, and stymied amyloid plaque production in the hippocampus, cortex, and brainstem by 6 months of age (see image below).
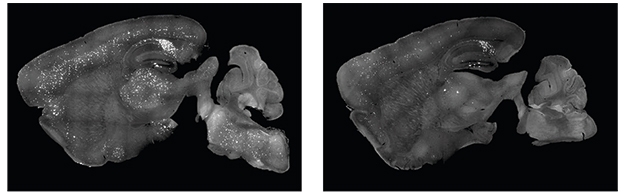
Peripheral Works, Too. 5xFAD mice intravenously injected with an AVV-PHP.eB-CRISPR targeting the APP Swedish mutation subsequently had fewer amyloid plaques (right) than controls (left). [Courtesy of Duan et al., Nature Biomedical Engineering, 2021.]
Systemic treatment also prevented microglial activation and dystrophic neurites in the hippocampus. In behavioral tests, the mice better remembered which arm of a Y-maze they had just explored, acclimated to an open-field as fast as wild-type, and spent less time on the open arms of an elevated plus maze test.
Of Mice and Marmosets: AAV for Better Brain Delivery
Achieving diffuse brain-specific delivery of an AAV virus that can efficiently edit genes remains a challenge for scientists. For example, AAV-PHP.eB also infiltrates the liver, where it can trigger an immune response from surveilling lymphocytes that reside there, or even damage the liver itself. “The success of gene therapies for CNS disorders will, to a large extent, depend on the development of AAV capsids with improved properties,” Ingelsson noted.
To minimize off-target effects, co-first authors David Goertsen, Nicholas Flytzanis, and Nick Goeden in Gradinaru’s lab tinkered with the AAV-PHP.eB capsid protein, VP3, to reduce its penchant to infiltrate the liver while maintaining, or even boosting, its preference for brain cells.
The scientists generated hundreds of thousands of AAV capsid sequences, each with a random combination of seven amino acids in a stretch of one surface-exposed loop of VP3. Then they infused this viral amalgam into peripheral veins of 8-week-old, wild-type mice, harvested the mice's organs, and sequenced DNA to identify which viruses had infected which tissue. They put the top 20 percent of capsids with high brain and low liver infectivity through the mouse infection screen again. Of the 39,000 capsid sequences that survived both rounds, the researchers selected 22 that most specifically infected CNS tissue.
One dubbed AAV.CAP-B10 stood out. It infected brain cells as well as AAV-PHP.eB but had negligible liver expression—50-fold lower than AAV-PHP.eB and more than 100-fold lower than AAV9. AAV.CAP-B10 infected four to five times fewer astrocytes and oligodendrocytes as did AAV-PHP.eB. To the authors, this indicated that AAV.CAP-B10 selectively infects neurons.
“The variants described by Goertsen et al. are truly of interest,” commented REGEMXBio's Danos. To the mind of Sergio Ferreira, Federal University of Rio de Janeiro, the role of glia cells in Alzheimer's pathogenesis raises the question of whether the capsid's specificity for neurons will prove to be an advantage or a drawback (full comment below).
Will this new virus behave the same way in people? That remains to be seen, but neither AAV-PHP.eB nor its cousin, AAV-PHP.B, infect nonhuman primate brain cells after systemic injection, dashing hope that these might be used for therapeutics (Matsuzaki et al., 2018). However, in two adult marmosets, a species of nonhuman primate, AAV.CAP-B10 infected four times as many neurons and 17-fold fewer liver cells than AAV9 or AAV-PHP.eB (see image below).
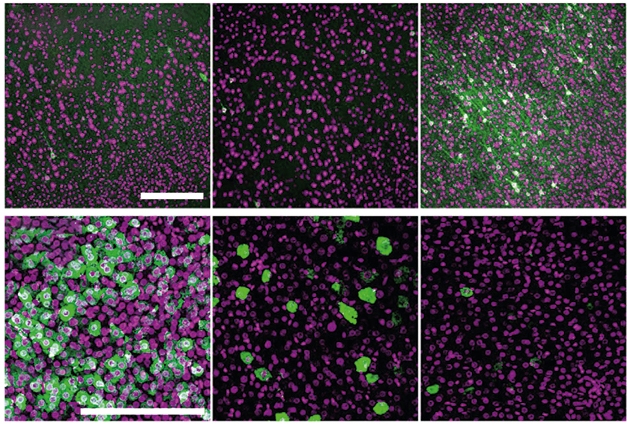
More Brain, Less Liver. In marmosets, AAV.CAP-B10 (right) encoding a hemagglutinin-labeled protein (green) better infected the cortex (top) than did AAV9 (left) or AAV-PHP.eB (middle), while its expression in the liver (bottom) was lower. Purple represents neurons (top) or nuclei (bottom). [Courtesy of Goertsen et al., Nature Neuroscience, 2021.]
Roy hopes that AAV capsids tested in nonhuman primates might work in people, given the successful translation from mice to marmosets. “This is important because testing efficacy of viral capsids in the human brain will be challenging in a clinical setting,” he wrote.
Ferreira wondered if AAV.CAP-B10 would improve the efficacy and specificity of the gene editing reported by Duan, Ye, and colleagues. Ip said her and Gradinaru's teams will develop intravenous APP gene editing in nonhuman primates using the CNS-optimized AAV capsids.—Chelsea Weidman Burke
References
Mutations Citations
News Citations
- In Mice, CRISPR-Based Alzheimer’s Therapies Inch Forward
- CRISPR Suppresses BACE1 and Plaques in Mice
- Gene Therapies Enter Trials for Many Brain Pathologies—What about AD?
Research Models Citations
Paper Citations
- Saraiva J, Nobre RJ, Pereira de Almeida L. Gene therapy for the CNS using AAVs: The impact of systemic delivery by AAV9. J Control Release. 2016 Nov 10;241:94-109. Epub 2016 Sep 13 PubMed.
- Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sánchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci. 2017 Aug;20(8):1172-1179. Epub 2017 Jun 26 PubMed.
- Matsuzaki Y, Konno A, Mochizuki R, Shinohara Y, Nitta K, Okada Y, Hirai H. Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci Lett. 2018 Feb 5;665:182-188. Epub 2017 Nov 24 PubMed.
Further Reading
News
Primary Papers
- Goertsen D, Flytzanis NC, Goeden N, Chuapoco MR, Cummins A, Chen Y, Fan Y, Zhang Q, Sharma J, Duan Y, Wang L, Feng G, Chen Y, Ip NY, Pickel J, Gradinaru V. AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat Neurosci. 2022 Jan;25(1):106-115. Epub 2021 Dec 9 PubMed.
- Duan Y, Ye T, Qu Z, Chen Y, Miranda A, Zhou X, Lok KC, Chen Y, Fu AK, Gradinaru V, Ip NY. Brain-wide Cas9-mediated cleavage of a gene causing familial Alzheimer's disease alleviates amyloid-related pathologies in mice. Nat Biomed Eng. 2021 Jul 26; PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
REGENXBIO
The best available AAV vectors used in the clinic today, AAV9 and AAVrh10, can only address a limited number of disease indications in the central nervous system. These are situations where local administration is possible (as in the striatum or in the hippocampus) or where a secreted therapeutic protein—an enzyme or an antibody—can be made by a minor fraction of modified neuronal cells and diffused throughout the CNS. High doses of vectors, potentially associated with toxicity, are needed to achieve efficacy. In cases where a genetic modification is needed in a large proportion of cells in an extended area such as the cortex, gene-transfer technologies are still suboptimal.
The paper by Goertsen et al. is a worthwhile read as it represents a milestone toward the development of gene-transfer vectors with an appropriate safety and efficacy profile for gene therapy of neurological diseases. Building on their previously described workflow, the Gradinaru lab describes novel AAV capsid variants with enhanced pharmacological and biodistribution properties in nonhuman primates (NHPs). The approach uses transgenic mice expressing Cre recombinase in various tissues and a lox-able vector design to identify variants with positive or negative tropism for a given target organ out of a complex library. Variants of interest are then independently characterized in Marmosets.…More
The group originally reported AAV9-derived variants AAV-PHP.B and AAV-PHB.eB with vastly improved performances, compared to the archetypal AAV9, for crossing the blood-brain barrier and targeting neural cells after intravenous administration. As generic tools for modifying brain cells in situ, these variants rapidly became a staples of neuroscience labs.
Several proofs of concept of brain-wide genetic modifications have been published, involving different gene-therapy modalities. The work described by Duan et al. uses a CRISPR Cas9 system and AAV-PHP.eB to knock out the AAPswe allele in transgenic mice with pathological features of Alzheimer's disease. The paper reports a surprisingly high level of gene editing throughout the brain (around 30 percent), a decrease in Aβ-associated pathologies, and an improvement in cognitive performance. Although the quantification and precise localization of gene editing would need to be confirmed and further established, this is an exciting example of what gene therapies for an intractable neurological condition could be. If, of course, all this can be demonstrably translated into the clinic—this where the latest variants may come into play.
The development of the original PHP.B variant toward the clinic has not been straightforward, essentially because their improved performances in rodents were not reproduced in NHPs where they behaved essentially as the parental AAV9. Here, Goertsen and colleagues selected new variants from a library built from PHP.eB, in which another variable surface loop of the capsid was randomized. Importantly, a negative selection of variants targeting the liver was also applied. The new variants obtained have enhanced tropism for the brain (markedly in mice, less so in NHPs) but interestingly, some of them are de-targeted from the liver.
This is the primary interest of these variants, because AAV capsids, including AAV-PHP.B, mostly end up in the liver once they access the circulation—this happens not only upon IV administration, but also with intra-CSF or intramuscular delivery. Orders of magnitude more genome copies are usually detected in the liver than at the target of interest, reducing bioavailability, potentially enhancing immune responses, and creating a genotoxic risk due to spurious integration of vector genomes in hepatocytes DNA.
The variants described by Goertsen et al. are truly of interest. Understanding how they work will be important: How do they reach the CNS, and what determined their cellular tropism there, i.e., neuron versus glia? Is Ly6B or a related protein the cellular receptor at play in BBB crossing? Is the original PHP.eB modification still needed, or would the new loop insertion work in an AAV9 background? What is the basis of escaping retention in the liver? Do they go to the spleen instead, as other de-targeted capsid mutants often do?
Almost certainly this team is already looking for third-generation variants that can avoid the dorsal root ganglion, a source of toxicity in preclinical studies with AAV9. As others in the field are also reporting new and interesting AAV capsids for CNS and other targets using comparable approaches, I expect to see a flurry of next-gen AAV capsids becoming available and empowering AAV gene therapy in the near future.
University of California, San Diego
The remarkable success of gene therapy for fatal spinal muscular atrophy has opened up a new paradigm for treating diseases afflicting the CNS, and it is difficult to imagine a future where gene-based therapies will not be translated to more common neurodegenerative diseases such as Alzheimer’s and Parkinson’s.
However, many neurodegenerative diseases will require diffuse delivery of genes to the large human brain, which remains a challenge. Moreover, though introduction of a single transgene into the CNS—such as that used in gene therapy for spinal muscular atrophy—is straightforward in principle, better tools are needed to harness the power of contemporary approaches such as CRISPR and base editing, which is more nuanced than a simple gene overexpression. The highlighted studies from the Gradinaru and Ip labs (Goertsen/Flytzanis/Goeden et al. and Duan/Ye et al.) advance both fronts.…More
Using a Cre-based screening and sequential engineering, Goertsen/Flytzanis/Goeden et al. identified AAV-based capsid variants that show remarkably widespread gene delivery in a nonhuman primate (marmoset) brain, while showing reduced targeting to non-CNS organs such as the liver. Moreover, one of the capsids (CAP-B10) showed high specificity for marmoset neurons, implying that we should be able to target different cellular populations in larger brains—for example neurons v/s glia—depending on what is needed for the specific disease. While the original screen was in mice, results translated well to marmosets, giving hope that capsids tested in nonhuman primates might also work in humans. This is important because testing efficacy of viral capsids in human brains will be challenging in a clinical setting. By achieving diffuse delivery in a larger brain, this work gets us one step closer to the goal of diffuse brain delivery in humans.
Duan/Ye et al. use a creative, CRISPR-based strategy to silence the “Swedish” mutation in the APP gene that causes a form of familial Alzheimer’s disease. Their AAV-based approach targets and degrades only the mutant APP allele, keeping the wild-type allele intact. They show that silencing the mutant allele in vivo with a single AAV injection alleviates neuropathology, behavior, and electrophysiologic alterations in a transgenic Alzheimer’s mouse model. They also use a single vector with a smaller Cas enzyme (SaCas9) that carried all the required CRISPR components in a single AAV, which is likely going to be the administration modality for most future CNS CRISPR therapies.
Indeed, an ongoing Phase I/I trial by Editas for an eye disease also uses the same SaCas9. Though applicable to a small fraction of Alzheimer’s patients carrying the familial Swedish mutation, this study opens the door for similar “personalized” CRISPR-based approaches for other Alzheimer’s mutations, which is potentially transformative for these unfortunate patients. Along with other gene-editing approaches that broadly target Alzheimer’s, for instance a strategy we are pursuing, CRISPR-based therapeutics offer some optimism against a disease where nothing seems to work (Sun et al., 2019).
References:
Sun J, Carlson-Stevermer J, Das U, Shen M, Delenclos M, Snead AM, Koo SY, Wang L, Qiao D, Loi J, Petersen AJ, Stockton M, Bhattacharyya A, Jones MV, Zhao X, McLean PJ, Sproul AA, Saha K, Roy S. CRISPR/Cas9 editing of APP C-terminus attenuates β-cleavage and promotes α-cleavage. Nat Commun. 2019 Jan 3;10(1):53. PubMed.
University of Toronto
The paper by Duan and colleagues describes the use and evaluation of CRISPR/Cas9 to disrupt the Swedish mutation of the APP gene. The authors have carefully tested this gene therapeutic strategy both via intracerebral delivery in 5xFAD and APP/PS1-transgenic mice and via AAV-mediated systemic delivery in 5xFAD mice. Interestingly, they found that both administration routes resulted in less brain pathology as compared to control mice. This work represents a clear step forward from our previous study, in which the same strategy was mainly evaluated in patient fibroblasts, and it is an important contribution to the development of CRISPR/Cas-based treatment strategies for dominantly inherited forms of neurodegenerative disorders (György et al., 2018). …More
The success of gene therapies for CNS disorders, based on CRISPR/Cas9 and other strategies, will to a large extent depend on the development of AAV capsids with improved properties. The PHP.eB subtype used by Duan et al. features widespread CNS transduction in rodents, but targets both neurons and glial cells. Moreover, it also transduces organs outside the brain—such as the liver—and does not effectively pass the blood-brain barrier in nonhuman primates. By deploying a synthetic library of sequence variants of a key region of the capsid, followed by a screen of such variants in cre-based transgenic mice, Goertsen and colleagues were able to identify PHP.eB versions with a preserved ability to cross the blood-brain barrier, and with tropism for neurons and avoidance for cell types outside the brain. One variant in particular with such properties, AAV.CAP-B10, was also found to efficiently transduce the marmoset brain upon peripheral administration.
This elegant piece of work from the researchers who previously generated PHP.eB represents an important step forward to enable future AAV-based systemic therapies for various brain disorders (Chan et al., 2017).
References:
György B, Lööv C, Zaborowski MP, Takeda S, Kleinstiver BP, Commins C, Kastanenka K, Mu D, Volak A, Giedraitis V, Lannfelt L, Maguire CA, Joung JK, Hyman BT, Breakefield XO, Ingelsson M. CRISPR/Cas9 Mediated Disruption of the Swedish APP Allele as a Therapeutic Approach for Early-Onset Alzheimer's Disease. Mol Ther Nucleic Acids. 2018 Jun 1;11:429-440. Epub 2018 Mar 16 PubMed.
Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sánchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci. 2017 Aug;20(8):1172-1179. Epub 2017 Jun 26 PubMed.
Federal University of Rio de Janeiro
Two complementary and joint studies from the groups of Viviana Gradinaru at Caltech and Nancy Ip at Hong Kong University highlight the potential of gene therapy for neurological disorders, in particular for familial Alzheimer’s disease (FAD). Combined, they take us one step closer to the day when targeted gene editing may allow resolution of genetic defects associated with FAD and other inherited neurological disorders.
In the first study, Yangyang Duan, from the Ip lab, and co-workers set out to investigate the potential benefits of AAV-mediated CRISPR-Cas9 delivery to the brain in two transgenic mouse models of AD, the 5xFAD and APP/PS1 mice. They began by designing an AAV9 vector simultaneously carrying Staphylococcus aureus Cas9 and one of two single-guide RNAs targeting two amino acid replacements in the amyloid precursor protein (APPswe) harbored in 5xFAD mice. After demonstrating that this AAV9 construct efficiently and specifically edited mutant APPswe (but not WT APP) in vitro, they injected it into the hippocampi of 3-month-old 5xFAD mice and examined their brains when they were 6 months old, an age at which they develop abundant brain amyloid deposits, gliosis, synapse impairment and loss. Interestingly, they found that disruption of APPswe reduced brain loads of both soluble and insoluble Aβ, reduced gliosis, and preserved synapse density (assessed by immunohistochemistry) and function (assessed by hippocampal LTP measurements) compared to mice that did not undergo allele editing. Similar beneficial actions were found in experiments with 9-month-old APP/PS1 mice, which already showed clear signs of neuropathology at the onset of the experiment.…More
Most interestingly, they then performed experiments employing an engineered AAV vector (AAV-PHP.eB) that is able to cross the blood-brain barrier. Using this approach, they showed that systemic delivery of the Cas9-sgRNA system resulted in efficient brain editing of APPswe in mice. In 5xFAD mice, this resulted in reductions in brain amyloid load and microgliosis and, remarkably, rescued some behavioral abnormalities presented by such mice.
While this study clearly demonstrates the potential of brain gene editing for monogenic neurological disorders, an issue that needs to be taken into consideration in the particular case of AD is that a small proportion (maybe as little as 3 percent) of total AD cases are of clear autosomal-dominant inheritance. This limits the applicability of the current approach to familial AD, meaning that chances appear slim that a similar approach could be developed that would comprise a universal therapy for the vast majority of sporadic AD cases. On the other hand, it should be emphasized that familial AD typically affects people at very young ages, often in their fifth or even fourth decades of life, and shows a very rapid progression. Once the results of genetic testing are complete, patients with mutations in APP, presenilin 1 (PS1), or presenilin 2 (PS2) essentially face an irrevocable sentence of AD. To these patients, therefore, the possibility of genetic targeting of disease-causing mutated alleles may represent a strong hope of evading that life sentence.
Another aspect that remains to be determined is whether the benefits of systemic AAV-mediated editing of brain genes are persistent or transient. It would be interesting to perform follow-up studies to determine whether the progression of AD neuropathology and functional deficits are stalled by AAV-Cas9 treatment as mice age further, or whether another round of treatment may be necessary. Finally, although Duan et al. have shown that working memory and anxiety-type behavior are improved by gene editing in 5xFAD mice, it would have been nice to see the potential beneficial effects in measures of hippocampus-dependent spatial orientation.
In the second study, co-lead authors Goertsen, Flytzanis, and Goeden (from the Gradinaru lab) and co-workers have paved new ground for the development of safe and effective viral-mediated gene therapies in the central nervous system. AAV-mediated gene therapy approaches have been under intensive investigation for CNS disorders for several years now. Although impressive cases of success have emerged, approaches are often hampered by challenges in vector delivery (e.g., requiring intrathecal or intracranial administration), low efficiency in transduction (often related to the need for parenchymal infusion, which limits diffusion of the viral construct), lack of cellular specificity, the possibility of systemic undesirable side-effects. Beginning with both naturally occurring serotypes and previously engineered AAV vectors, and using methods of directed evolution and AAV engineering, the current study generated a very large number of changes in AAV capsid, and screened them for optimal CNS targeting and minimal targeting of peripheral organs in mice.
Among thousands of capsids screened, a few were quite effective in targeting the brain while at the same time avoiding peripheral organs when administered systemically in mice. One of them, AAV.CAP-B10, was particularly effective in transducing brain cells while exhibiting virtually no transduction in the liver (which is highly targeted by systemic administration of previously used AAV vectors). Of particular interest was the comparison between AAV.CAP-B10 and AAV-PHP.eB, which was previously developed to cross the blood-brain barrier and was used in the study discussed above. While systemic administration of both AAV.CAP-B10 and AAV-PHP.eB in mice effectively targeted the brain, the latter significantly targeted the liver. In addition, greater cellular specificity was also achieved with AAV-CAP-B10, which largely transduced neurons (and not glia).
Goertsen et al. further showed that systemic administration of AAV.CAP-B10 efficiently targeted the brain of marmosets, a relevant finding in terms of the translational potential of this approach in primates. Overall, AAV.CAP-B10 appears as a powerful tool to expand the repertoire of viral-mediated therapeutic approaches in the central nervous system while minimizing peripheral side-effects. It would be interesting to determine, for example, if use of AAV.CAP-B10 could improve efficiency of the Cas9-mediated gene-disruption approach used in the study by Duan et al., while at the same time abrogating peripheral targeting. On the other hand, in light of the increasing recognition of the role of glial cells (notably microglia and astrocytes) in the pathogenesis of AD and of the growing number of glial proteins that are risk factors for AD, it remains to be determined whether enhanced specificity toward neurons would represent an advantage or a drawback of AAV.CAP-B10.
Make a Comment
To make a comment you must login or register.