Double Spiral Sets TDP-43 Apart from Other Amyloids
Quick Links
When researchers used cryo-electron microscopy to zoom in on TDP-43 filaments taken from the brains of two people with ALS/FTD, what they found looked like something viewed through the Hubble telescope. According to a study published December 8 in Nature, researchers led by Benjamin Ryskeldi-Falcon of the MRC Laboratory of Molecular Biology, Cambridge, U.K., saw a galaxy-like, double-spiral structure at the core of the filaments. Structures from different cortical regions were the same. They were filaments consisting of stacks of TDP-43 molecules, each wrapped into a double-spiral fold unlike any amyloidogenic protein observed so far.
- Cyro-EM resolves atomic structures of TDP-43 filaments from two people with ALS/FTD.
- Protofilaments' core forms a unique double-spiral fold.
- The structure distinguishes brain filaments from those made in vitro.
The structure bore no resemblance to any TDP-43 filament formed in vitro. This may help explain why TDP-43 rebuffs binding by PET tracers and dyes, and may illuminate neuropathological connections among disorders marked by TDP-43 pathology.
“This exciting work presents the first atomic structures of pathological amyloid fibrils of TDP-43 extracted from the brains of two patients who suffered from ALS with FTLD,” Dan Li of Shanghai Jiao Tong University, China, wrote to Alzforum. “Given the important role of TDP-43 aggregation in ALS, FTLD, and some other neurodegenerative diseases, this work provides an important starting point to understand the structure-pathology relationship of TDP-43 aggregation in diseases.”
Manuela Neumann of the German Center for Neurodegenerative Diseases in Tübingen co-discovered TDP-43. To her mind, the lack of agreement with filament structures from in vitro cryo-EM underscores the importance of postmortem tissue for structural studies (full comment below).
An RNA-binding protein, TDP-43 spends most of its time in the nucleus. However, in most cases of ALS and about half of FTD cases, the protein aggregates, and can wind up marooned in the cytoplasm of neurons and glial cells. TDP-43 pathology also has been spotted in people with AD and other diseases, and has led scientists to propose a distinct neuropathological entity called limbic-predominant, age-related TDP-43 encephalopathy (LATE) (May 2019 news).
How the structure of TDP-43 aggregates relates to these diverse clinical and neuropathological manifestations remains unclear. Efforts to analyze the structure of TDP-43 filaments have thus far been limited to those formed in vitro by portions or all of the low-complexity domain of the protein (Cao et al., 2019; Li et al., 2021).
First author Diana Arseni and colleagues wanted to resolve the structure of TDP-43 filaments from within the brain. They obtained tissue samples from two people who had died with advanced ALS, and who also had FTD. Whether extracted from the motor or the frontal cortex, TDP-43 aggregates comprised both full-length and C-terminal fragments. Immuno-electron microscopy revealed that the TDP-43 aggregates consisted of helical filaments, with a width of 10 to 15 nm.
To gaze at the heart of the filament at the atomic level, the researchers turned to cryo-EM. They extracted sufficient TDP-43 for this purpose from the frontal and motor cortex samples of one donor, and from the frontal cortex of the other. Before subjecting the samples to cryo-EM, the researchers stripped away the nebulous coat from the filaments with pronase, leaving intact only their core. At a resolution of 2.6Å, a single-filament structure emerged from all three samples: one stack of TDP-43 molecules, spaced 4.8Å apart to form a filament with a right-handed helical twist of 1.4 degrees.
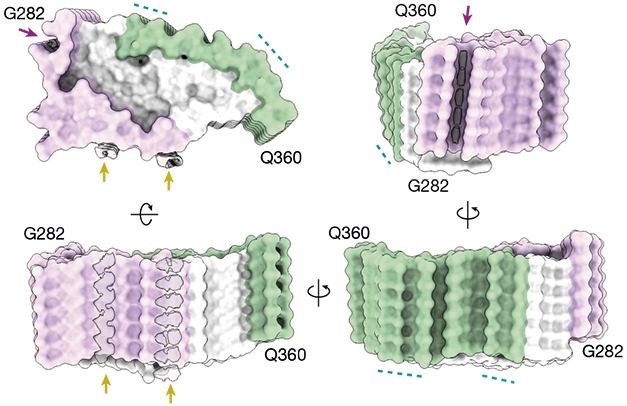
Stacked Spiral. Viewed from various angles, TDP-43 filaments comprised a single stack of TDP-43 molecules, each wrapped into a double-spiral fold divided into a hydrophobic core (white), glycine-rich region (purple), and glutamine/asparagine-rich region (green). The filament formed with a right-handed helical twist. [Courtesy of Arseni et al., Nature, 2021.]
Perpendicular to the helical axis, each TDP-43 molecule wrapped itself into a double spiral. This type of structure hasn’t been seen for other amyloidogenic proteins. Instead, Falcon said, it bears an uncanny resemblance to something otherworldly: massive spiral galaxies.
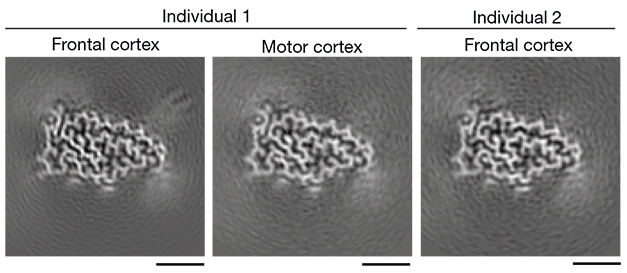
Space-y Similarity. In both patients and in two brain regions, TDP-43 adopted the same distinctive double-spiral fold. [Courtesy of Arseni et al., Nature, 2021.]
Formed by 79 residues of the low-complexity domain of TDP-43, the fold can be divided into three domains based on amino acid composition: a glycine-rich region toward the N-terminus, a hydrophobic region in the middle, and a region rich in glutamine and asparagine toward the C-terminus. The hydrophobic region formed the spiral’s core, which was snugly wrapped by the glycine- and glutamine/asparagine-rich regions of the spiral arms.
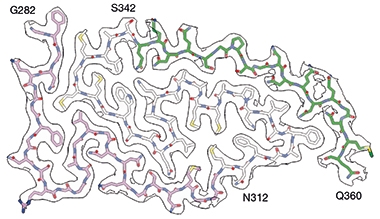
Spiral Hug. A hydrophobic region (white residues) formed the core of TDP-43’s double spiral, held tightly by a glycine-rich (purple) and a glutamine/asparagine-rich arm (green). [Courtesy of Arseni et al., Nature, 2021.]
The double spiral sets TDP-43 filaments apart from known amyloids. The structure has none of the long β-sheets that forge strong contacts between protofilaments in Aβ, tau, and α-synuclein filaments. Each TDP-43 spiral consisted of 10 tiny β-sheets, only two of which topped three amino acids in length, and none made contact with other spirals in the filaments. Filaments of other heterogenous ribonucleoproteins (hnRNPs), including FUS, hnRNPA1, and hnRNPA2, also have been shown to accommodate multiple small β-sheets, at least in vitro. However, these differ from TDP-43 filaments in that they are enriched with tyrosine residues and lack a hydrophobic core. To Falcon's mind, the latter likely explains why TDP-43 filaments remain stable at 65oC, while other hnRNP filaments fall apart.
Falcon cautioned against assuming hnRNP filaments formed in vitro would be representative of those in the brain. In fact, the ALS/FTD TDP-43 filament was entirely distinct from those formed, in vitro, from segments of the protein or by using harsh conditions, such as low pH. Some of those synthetic fibrils have dagger- and R-shaped folds, or an entirely different shape, as recently obtained by Witold Surewicz and colleagues at Case Western Reserve University, Cleveland (see Li et al., 2021, and image below).
Why is that? In the brain, TDP-43 interacts with multiple proteins as well as RNA. It undergoes phase separation, joining up with membraneless organelles such as stress granules. These manifold interactions likely dictate how the protein folds in its natural environment, Falcon said.
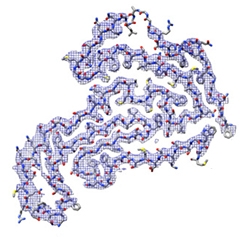
Synthetic TDP43. TDP filaments formed in a test tube take a different shape from those extracted from human brain. [Courtesy of Li et al., Nature Communications, 2021.]
Li noted that post-translational modifications and ligands may play an important role in fibril polymorph selection, as well. “Indeed, several unidentified densities were observed adjacent to the TDP-43 fibril surface. Identification of these unknown ligands may provide a clue for fibril polymorph selection,” he wrote. Unidentified entities have been detected by cryo-EM of tau and synuclein fibrils, as well (Oct 2021 news; Mar 2020 conference news).
Neumann noted that the reported differences to Aβ, tau, or synuclein filaments might explain why TDP-43 aggregates poorly bind amyloid dyes, such as Thioflavin S. Falcon and colleagues found that unlike other amyloids, TDP-43 filaments have no deep grooves. Rather, the glycine- and glutamine/asparagine-rich spiral arms form smooth surfaces with only a single groove. This could explain why PET ligands, too, which nestle into the pockets of other amyloid filaments, can't stick to aggregated TDP-43. A new crop of compounds may be needed to bind to TDP-43’s uniquely smooth surface.
The samples used in this study came from people whose ALS/FTD featured type B TDP-43 pathology, which is characterized by diffuse or granular neuronal cytoplasmic inclusions without dystrophic neurites. Other neuropathological subtypes of TDP-43 exist in people with different forms of FTD, as well as in LATE, and Falcon plans to use cryo-EM to solve their structures as well. The findings could help draw connections and distinctions between different disorders marked by TDP-43 pathology, he said.
Neumann added that it will be crucial to learn if, and how, TDP-43 filaments differ among the various subtypes of FTLD-TDP, and between different brain regions such as the spinal cord and brainstem.—Jessica Shugart
References
News Citations
- Introducing LATE—A Common TDP-43 Proteinopathy that Strikes After 80
- Flock of New Folds Fills in Tauopathy Family Tree
- Behold the First Human α-Synuclein CryoEM Fibril Structure
Paper Citations
- Cao Q, Boyer DR, Sawaya MR, Ge P, Eisenberg DS. Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat Struct Mol Biol. 2019 Jul;26(7):619-627. Epub 2019 Jun 24 PubMed.
- Li Q, Babinchak WM, Surewicz WK. Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43. Nat Commun. 2021 Mar 12;12(1):1620. PubMed.
Further Reading
Primary Papers
- Arseni D, Hasegawa M, Murzin AG, Kametani F, Arai M, Yoshida M, Ryskeldi-Falcon B. Structure of pathological TDP-43 filaments from ALS with FTLD. Nature. 2022 Jan;601(7891):139-143. Epub 2021 Dec 8 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
University of Tübingen and DZNE AG Neumann
I am very pleased to see cyroEM data on TDP-43 filaments finally emerging, and I would like to congratulate the authors. In the study, the authors focused on the analysis of cortical TDP-43 aggregates isolated from human postmortem tissue from two ALS/FTLD type B TDP cases. They describe, for the first time, the structure of the pronase-resistant core of TDP-43 filaments.
The reported evident structural differences from filaments formed by other proteins involved in neurodegenerative diseases, such as tau or α-synuclein, provide a good explanation for the well-described distinct behavior of TDP-43 aggregates with respect to binding to amyloid dyes, such as Thioflavin S. Also of particular note is the lack of agreement with reported TDP-43 filament structures from in vitro cryoEM studies, which supports the essential importance of postmortem tissue analysis for these kinds of studies.
In the future, it will be crucial to learn if and how the structure of TDP-43 filaments differs among the various well-recognized subtypes of FTLD-TDP. Moreover, given the well-recognized difference in the composition of TDP-43 inclusions in the spinal cord/brainstem compared to those in cortical CNS regions in terms of abundance of full-length versus C-terminal fragments, it will be also very interesting and important to see whether the structure of the filaments will be also consistent across these different CNS regions.
Shanghai Jiao Tong University
This exciting work presents the first atomic structures of pathological amyloid fibrils of TDP-43 extracted from the brains of two patients who suffered from ALS with FTLD. The TDP-43 fibrils exhibit a novel compact double-spiral-shaped fold. Given the important role of TDP-43 aggregation in ALS, FTLD and some other neurodegenerative diseases, this work provides an important starting point to understand the structure-pathology relationship of TDP-43 aggregation in diseases.
Interestingly, unlike α-synuclein and Aβ fibrils, both of which show structural similarity between in vitro-prepared and brain-derived fibrils to some extent (Schweighauser et al., 2020; Li et al., 2018; Li et al., 2018; Yang et al., 2021; Schütz et al., 2015), TDP-43 fibril is completely distinct from those prepared in vitro in terms of helical chirality, secondary structure, and the size of fibril core. This observation suggests a high degree of conformational plasticity of TDP-43 in forming fibrils which may be due to the low-complexity nature of its primary sequence. Indeed, different TDP-43 strains were previously identified in the brains of different FTLD-TDP subtypes with distinct pathological properties (Porta et al., 2021). It will be interesting to see whether TDP-43 adopts different atomic structures in fibrils of other subtypes.
If so, a question will be how the double-spiral-shaped fold of TDP-43 fibril is determined in the brain of a patient with ALS with FTLD? Previous studies showed that PTM and ligands may play an important role in fibril polymorph selection (Li and Liu, 2021; Arakhamia et al., 2020). Indeed, several unidentified densities were observed adjacent to the TDP-43 fibril surface formed by Q286, R293 and A315. Identification of these unknown ligands may provide a clue for fibril polymorph selection.
Given the resolution revolution of cryo-EM, more and more amyloid fibril structures assembled in vitro or extracted from brains were determined in the past few years. However, a key question remains to be answered: How do amyloid fibril structures explain their pathologies? The connection between fibril structure and pathology needs to be established.
References:
Schweighauser M, Shi Y, Tarutani A, Kametani F, Murzin AG, Ghetti B, Matsubara T, Tomita T, Ando T, Hasegawa K, Murayama S, Yoshida M, Hasegawa M, Scheres SH, Goedert M. Structures of α-synuclein filaments from multiple system atrophy. Nature. 2020 Sep;585(7825):464-469. Epub 2020 May 27 PubMed.
Li Y, Zhao C, Luo F, Liu Z, Gui X, Luo Z, Zhang X, Li D, Liu C, Li X. Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res. 2018 Sep;28(9):897-903. Epub 2018 Jul 31 PubMed.
Li B, Ge P, Murray KA, Sheth P, Zhang M, Nair G, Sawaya MR, Shin WS, Boyer DR, Ye S, Eisenberg DS, Zhou ZH, Jiang L. Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat Commun. 2018 Sep 6;9(1):3609. PubMed.
Yang Y, Arseni D, Zhang W, Huang M, Lövestam S, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Ryskeldi-Falcon B, Scheres SH, Goedert M. Cryo-EM structures of amyloid-β 42 filaments from human brains. Science. 2022 Jan 14;375(6577):167-172. Epub 2022 Jan 13 PubMed.
Schütz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Güntert P, Böckmann A, Glockshuber R, Meier BH. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl. 2015 Jan 2;54(1):331-5. Epub 2014 Nov 13 PubMed.
Porta S, Xu Y, Lehr T, Zhang B, Meymand E, Olufemi M, Stieber A, Lee EB, Trojanowski JQ, Lee VM. Distinct brain-derived TDP-43 strains from FTLD-TDP subtypes induce diverse morphological TDP-43 aggregates and spreading patterns in vitro and in vivo. Neuropathol Appl Neurobiol. 2021 May 10; PubMed.
Li D, Liu C. Hierarchical chemical determination of amyloid polymorphs in neurodegenerative disease. Nat Chem Biol. 2021 Mar;17(3):237-245. Epub 2021 Jan 11 PubMed.
Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, Seyfried NT, Petrucelli L, Fitzpatrick AW. Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell. 2020 Feb 20;180(4):633-644.e12. Epub 2020 Feb 6 PubMed.
Make a Comment
To make a comment you must login or register.