Shape Your Microbiome. You’ll Live Longer, Scientists Say
Quick Links
There’s a new reason to “trust your gut,” according to a study published February 18 in Nature Metabolism. Researchers led by Nathan Price at the Institute for Systems Biology, a nonprofit biomedical research organization in Seattle, found that the trillions of bacteria, viruses, fungi, and other microorganisms that make up the human gut microbiome change as we age. Curiously, they report, the more idiosyncratic the digestive system’s microbiome becomes to a given person, the longer that person lives, and the better his or her health.
- Gut flora changes as people get older.
- A person’s microbiome becomes more individualized starting in the 40s and 50s.
- In people over 84, this uniqueness appears linked to a longer lifespan.
“This is a fascinating study of gut microbiome in older adulthood,” wrote Barbara Bendlin from the University of Wisconsin, Madison. “While the investigators did not look at brain health or cognitive outcomes, it’s interesting to see that they found that healthy aging was accompanied by gut microbiomes that became increasingly more unique to each person starting in middle age. This type of divergence is also observed in brain aging.” (Comment below.)
Past studies have shown that the gut microbiome undergoes rapid changes in the first three years of life, followed by a longer period of relative stability, then more change once again in later years (Yatsunenko et al., 2012; O’Toole and Jeffery, 2015). Research has also found that centenarians have fewer of the gut microbes commonly seen in younger, healthy people. Instead, they live with an increasingly rarefied microbiota (Kim et al., 2019). This suggests that gut microbiomes become increasingly personalized as people get older, but little is known about how these gut profiles affect the aging process or longevity.
To find out, first author Tomasz Wilmanski and colleagues analyzed gut microbiomes, personal traits, and clinical data from more than 9,000 people 18 to 101 years old. They came from three independent cohorts. One was a group of 3,653 people aged 18 to 87 who had signed up with Arivale, a now-defunct scientific wellness company co-founded by systems biology pioneer Leroy Hood and Price. Arivale provided personalized wellness coaching by collecting and analyzing data on participants’ genomes and other systems, including their gut microbiomes. Hood founded the Institute for Systems Biology.
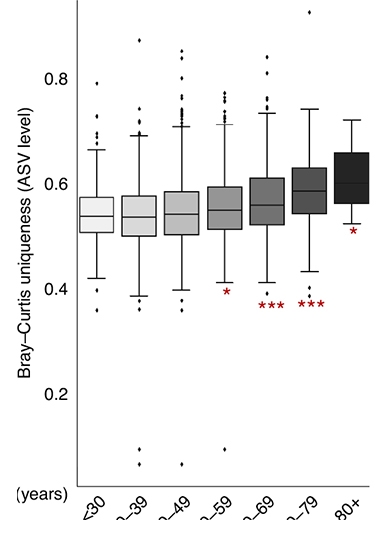
Ripe Old Age? In the Arivale cohort, the older participants got, the more their gut microbiomes matured in terms of amplicon sequence variants. [Courtesy of Wilmanski et al., Nature Metabolism, 2021.]
Wilmanski calculated the microbiomes’ Bray-Curtis uniqueness; this is a measure of how dissimilar the microbiomes are in one person compared to another in terms of the type of bugs present and their abundance. He found that people’s gut microbiomes started to become more distinctive at the genus level during their 40s and 50s. In the 50s, 60s, and beyond, the gut became more divergent at the amplicon sequence level, a term that denotes individual organisms. Amplicons are basically sequenced stretches of DNA.
Along with age, Wilmanski also looked at other variables likely influence one’s microbiome, including body mass index, alcohol consumption, and prescription medicine use. Most of them didn’t survive adjustment for age, except for lipid markers. People with more individualized microbiome signatures had lower low-density lipoprotein-cholesterol and triglyceride, and higher vitamin D levels, all known markers of health.
Are there health implications to microbiome individuation in late life? Wilmanski looked at the Osteoporotic Fractures in Men (MrOs), a longitudinal observational cohort of 907 men aged 78 to 98. Calculating their Bray-Curtis uniqueness, he found that the most “unique” microbiomes had fewer of the two commonly seen gut bacteria Bacteroides and Prevotella. This drop was linked to better health and longevity. On the other hand, people aged 85 and older whose intestines retained high levels of bugs such as Bacteroides and Prevotella, and had less individuated microbiomes, were less likely to survive during the next four years (see image below).
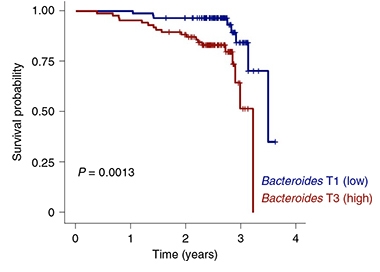
Less Is More. Participants aged 85 and older with more Bacteroides in their guts (blue) died younger than those with fewer. [Courtesy Wilmanski et al., Nature Metabolism, 2021.]
Previously, the authors found that certain plasma metabolites predict the diversity of a person’s gut microbiome (Wilmanski et al, 2019). To expand on this, Wilmanski tested 653 metabolites in the Arivale cohort to see if any of them reflected the gut microbiome compositions that develop with aging. Derivatives of tryptophan and phenylalanine, including indoles and phenylacetylglutamine, correlated with microbiome uniqueness. Other studies have reported more activation of the pathways that lead to tryptophan and phenylalanine metabolism in the gut microbiomes of centenarians than in younger controls (Rampelli et al., 2013; Collino et al., 2013). Additionally, indole mediates inflammation and extends survival in animal models (Krishnan et al., 2018; Sonowal et al., 2017).
These findings were confirmed among 4,575 participants from the American Gut Project, the largest publicly available human gut bacterial dataset (McDonald et al., 2018).
The study confirms that aging may bring gradual shifts in gut metabolic capacity, and supports the idea that blood metabolites reflect the function and individuation of the microbiome, wrote the authors. “You can often get more information about the health state of your microbiome from looking at the metabolites that you find in the blood, as opposed to the microbial species themselves, which vary from person to person and place to place,” Price told Alzforum. Researchers in this young field are struggling to make sense of the diversity of the microbiome and how it affects health and disease.
The authors see their study as a step forward in understanding how the human gut impacts health. “Basically, if you stay healthy, your microbiome will evolve along with you,” said Price. “It changes and it optimizes and becomes increasingly unique. That doesn’t happen if you’re not healthy.”
But what makes the gut microbiome healthy? Wilmanski and Price don’t know. “We are still in the early stages of really understanding in depth how to steer a microbiome,” said Price. “It’s not so hard to define what is unhealthy in the microbiome, but it’s harder to find what is good.”—Helen Santoro
References
Paper Citations
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012 May 9;486(7402):222-7. PubMed.
- O'Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015 Dec 4;350(6265):1214-5. PubMed.
- Kim BS, Choi CW, Shin H, Jin SP, Bae JS, Han M, Seo EY, Chun J, Chung JH. Comparison of the Gut Microbiota of Centenarians in Longevity Villages of South Korea with Those of Other Age Groups. J Microbiol Biotechnol. 2019 Mar 28;29(3):429-440. PubMed.
- Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, Lovejoy J, Omenn GS, Hood L, Gibbons SM, Price ND. Blood metabolome predicts gut microbiome α-diversity in humans. Nat Biotechnol. 2019 Oct;37(10):1217-1228. Epub 2019 Sep 2 PubMed.
- Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, O'Toole PW, Brigidi P. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY). 2013 Dec;5(12):902-12. PubMed.
- Collino S, Montoliu I, Martin FP, Scherer M, Mari D, Salvioli S, Bucci L, Ostan R, Monti D, Biagi E, Brigidi P, Franceschi C, Rezzi S. Metabolic signatures of extreme longevity in northern Italian centenarians reveal a complex remodeling of lipids, amino acids, and gut microbiota metabolism. PLoS One. 2013;8(3):e56564. Epub 2013 Mar 6 PubMed.
- Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, Yarmush ML, Alaniz RC, Jayaraman A, Lee K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018 Apr 24;23(4):1099-1111. PubMed.
- Sonowal R, Swimm A, Sahoo A, Luo L, Matsunaga Y, Wu Z, Bhingarde JA, Ejzak EA, Ranawade A, Qadota H, Powell DN, Capaldo CT, Flacker JM, Jones RM, Benian GM, Kalman D. Indoles from commensal bacteria extend healthspan. Proc Natl Acad Sci U S A. 2017 Sep 5;114(36):E7506-E7515. Epub 2017 Aug 21 PubMed.
- McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, DeRight Goldasich L, Dorrestein PC, Dunn RR, Fahimipour AK, Gaffney J, Gilbert JA, Gogul G, Green JL, Hugenholtz P, Humphrey G, Huttenhower C, Jackson MA, Janssen S, Jeste DV, Jiang L, Kelley ST, Knights D, Kosciolek T, Ladau J, Leach J, Marotz C, Meleshko D, Melnik AV, Metcalf JL, Mohimani H, Montassier E, Navas-Molina J, Nguyen TT, Peddada S, Pevzner P, Pollard KS, Rahnavard G, Robbins-Pianka A, Sangwan N, Shorenstein J, Smarr L, Song SJ, Spector T, Swafford AD, Thackray VG, Thompson LR, Tripathi A, Vázquez-Baeza Y, Vrbanac A, Wischmeyer P, Wolfe E, Zhu Q, American Gut Consortium, Knight R. American Gut: an Open Platform for Citizen Science Microbiome Research. mSystems. 2018 May-Jun;3(3) Epub 2018 May 15 PubMed.
External Citations
Further Reading
Primary Papers
- Wilmanski T, Diener C, Rappaport N, Patwardhan S, Wiedrick J, Lapidus J, Earls JC, Zimmer A, Glusman G, Robinson M, Yurkovich JT, Kado DM, Cauley JA, Zmuda J, Lane NE, Magis AT, Lovejoy JC, Hood L, Gibbons SM, Orwoll ES, Price ND. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat Metab. 2021 Feb;3(2):274-286. Epub 2021 Feb 18 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
University of Wisonsin
This is a fascinating study of gut microbiome in older adulthood. While the investigators did not look at brain health or cognitive outcomes, it is interesting that they found that healthy aging was accompanied by gut microbiomes that became increasingly more unique to each person starting in middle-age.
This type of divergence in trajectories of aging is something that is also observed in the brain. If you examine groups of older adults they start to “spread out,” with many individuals maintaining good cognitive health, while others showing more decline, and still others progressing to dementia. It would be so interesting to see whether the gut microbiome effects observed in this study track with brain health into older age in longitudinal studies.…More
What we don’t know from this study is why the gut microbiome is becoming more unique in older age, particularly among the healthy older adults. Does the gut drive patterns of health and disease, or does the gut microbiome respond to age-related health threats to actually become more supportive of better outcomes? Perhaps the gut microbiomes of some individuals are more readily able to respond to the effects of aging than the gut microbiomes of others. Conversely, the results may reflect the environments and life experiences that contribute to both successful aging and gut composition.
Additional studies, particularly those that involve longitudinal sample collection, will be needed to address these questions. Kudos to Dr. Rima Kaddurah-Daouk and colleagues for pursuing their large multisite longitudinal studies on gut microbiome, which are critically needed to better determine the link between gut, brain aging, and development of neurodegenerative disease (Alzheimer’s Gut Microbiome Project).
OHSU
This study by Wilmanski et al. provides strong additional support for the idea that alterations in, distinct signatures of, and taxonomic diversity and abundance within the gut microbiome play important roles in physiological versus pathological aging, and even in survival in the oldest-old (over 80 years).
Using three large, independent cohorts, the authors show that compositional uniqueness is associated with microbial amino acid metabolites in the plasma, including phenylalanine/tyrosine and tryptophan, which have been implicated previously in immune function, aging, and longevity. Healthy aging associated with a drift toward a more unique compositional state, including a depletion of Bacteroides. In contrast, unhealthy aging associated with a decline in the Lachnoclostridium and Rumminococace genera. The drift toward an increasingly unique gut microbiome composition started between 40 and 50 years of age at the genus level, and between 50 and 60 years at the amplicon sequence variant level, and continued to increase with every decade.…More
Some of the identified taxa that positively associated with uniqueness, both beneficial (Christensenellaceae) and potentially pathogenic (Methanobrevibacter and Desulfibrio), were previously implicated in human longevity. Based on health heterogenicity, the authors could stratify MrOS study participants based on four measures: medication use, self-perceived health (excellent versus less than excellent), life-space score (LSC) (how often an individual leaves his or her room, house, or neighborhood) and walking speed. For both the LSC and walking speed, the authors compared the top tertile with the bottom two. The correlation between uniqueness and age remained significant in highly medicated (more than eight medications) healthy individuals and was independent of sex. Prescription medication use and alcohol consumption significantly associated with uniqueness but after adjusting for age, only lipid markers remained significantly associated with gut microbiome uniqueness. The direction of association indicated healthier metabolic and lipid profiles, including lower low-density lipoprotein-cholesterol, higher vitamin D and lower triglycerides, in individuals with more unique microbiomes.
These data are consistent with increasing evidence supporting for a role for alterations in the gut microbiome in brain function. The gut microbiome can communicate with the brain and affect neurobiology and behavioral phenotypes, including stress-related behaviors, anxiety, and depression (Foster et al., 2013; Allen et al., 2017; Kelly et al., 2015; Lynch and Hsiao, 2019; Vuong et al., 2017; Sudo et al., 2004). For example, gut microbiota regulate motor impairments and neuroinflammation in an α-synuclein-based Parkinson’s disease mouse model (Sampson et al., 2016), and we have shown that the effects of the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) on cognitive performance may be, at least in part, mediated by the gut microbiome (Kundu et al., 2021). MPTP affected the diversity of the gut microbiome and there were significant associations between microbiome α-diversity and sensorimotor performance, as well as microbiome composition and fear learning.
The gut microbiome might be important in Alzheimer’s disease as well (Magnusson et al., 2015). Microbiome perturbations using an antibiotic cocktail reduced Aβ pathology, astrogliosis, and microglial morphology in male mice overexpressing both human amyloid precursor protein (hAPP) with the Swedish mutation and human presenilin 1, and transplants of fecal microbiota of genotype- and age-matched male mice partially rekindled Aβ pathology and microglial morphology (Dodiya et al., 2019).
Our data in hAPP knock-in (KI) mice containing the Swedish and Iberian mutations (AppNL-F), or those variants and the Arctic mutation (AppNL-G-F), further support that alterations in the gut microbiome composition might contribute to AD (Kundu et al., 2021). Behavioral and cognitive performance in 6-month-old AppNL-F, AppNL-G-F, and C57BL/6J wild-type (WT) mice were associated with the gut microbiome. Genotype modulated these relationships, which were also test-dependent, as evidenced by our β diversity analysis, revealing baseline differences in the microbiome based on genotype. For example, the biodiversity of the gut microbiome negatively associated with the amount of time WT mice spent exploring a novel object, which is an indicator of object-recognition memory. However, the biodiversity within AppNL-F and AppNL-G-F mice manifested positive associations with this same measure. The composition of the gut microbiome manifested striking differences as a function of genotype, with all three genotypes eliciting distinct microbiome compositions. Moreover, the association between the composition of the microbiome and the time a mouse spent exploring a novel object differed in a genotype-dependent manner. Accordingly, the APP genotype also affected the relationship between the relative abundance of specific phylotypes in the gut and various behavioral and cognitive measures.
These genotype-dependent associations involved members of the Lachnospiraceae and Ruminococcaceae families, which in our prior data in the MPTP PD model described above and in B6D2F1 mice exposed to simulated space radiation were linked to behavior and cognitive performance (Magnusson et al., 2015; Raber et al. 2020; Torres et al., 2018). Intriguingly, the Ruminococcaceae families also came up in the current human study. That does not mean that the direction of gut microbiome changes in humans and animal models and the direction of the relationships between gut microbiome and behavioral and cognitive performance measures or other health-related measures necessarily go in the same direction across all studies. Our mouse data indicate that the directions of these relationships are genotype-dependent. In addition, as discussed by the authors, there might be differences in aging gut dynamics in distinct human populations (for example, healthy community-dwelling elderly versus fragile long-term-care residents) and in the resilience of individuals to detrimental effects of the gut microbiome on plasma metabolites.
Epigenetic changes in the hippocampus might be involved in mediating the effects of the gut microbiome on the brain. In a subset of female mice, we investigated whether alterations in hippocampal DNA methylation were associated with the gut microbiome. An integrated gut microbiome/hippocampal DNA methylation analysis revealed a positive relationship between amplicon sequence variants within the Lachnospiraceae family and methylation at the Apoe gene (Kundu et al., 2021). These microbes may elicit an effect on AD-relevant behavioral and cognitive performance via epigenetic changes in AD-susceptibility genes in neural tissue. Alternatively, epigenetic changes might elicit alterations in intestinal physiology that affect the growth of these taxa in the gut.
As sex-dependent effects are seen in gut microbiome studies in animal models and the MrOS cohort only includes men, it was important that the correlation between uniqueness and age remained independent of sex. A potential limitation of this study is that alcohol use might be an important contributor to the results, because alcohol use majorly affects the gut microbiome and therefore masks other effects (Leviatan and Segal, 2020).
The age window at which the gut microbiome becomes more unique in people with a healthy trajectory and distinct from those with a less-healthy trajectory suggests that alterations in the gut microbiome might be important predictive biomarkers of healthy and less-healthy aging. The gut microbiome might be important to consider for predicting treatment responses in patients with neurodegenerative conditions as well. For example, the gut microbiome profile is critical in the response of patients with metastatic melanoma to checkpoint inhibitor immunotherapy (Limeta et al., 2020) and some refractory patients respond following fecal microbiota transplantation (No authors listed, Cancer Discov., 2021). Clearly, increased efforts are warranted to study the role of the gut microbiome in brain function under physiological and pathological conditions in humans and animal models.
References:
Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013 May;36(5):305-12. Epub 2013 Feb 4 PubMed.
Allen AP, Dinan TG, Clarke G, Cryan JF. A psychology of the human brain-gut-microbiome axis. Soc Personal Psychol Compass. 2017 Apr;11(4):e12309. Epub 2017 Apr 18 PubMed.
Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front Cell Neurosci. 2015;9:392. Epub 2015 Oct 14 PubMed.
Lynch JB, Hsiao EY. Microbiomes as sources of emergent host phenotypes. Science. 2019 Sep 27;365(6460):1405-1409. PubMed.
Vuong HE, Yano JM, Fung TC, Hsiao EY. The Microbiome and Host Behavior. Annu Rev Neurosci. 2017 Jul 25;40:21-49. Epub 2017 Mar 8 PubMed.
Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004 Jul 1;558(Pt 1):263-75. Epub 2004 May 7 PubMed.
Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson's Disease. Cell. 2016 Dec 1;167(6):1469-1480.e12. PubMed.
Dodiya HB, Kuntz T, Shaik SM, Baufeld C, Leibowitz J, Zhang X, Gottel N, Zhang X, Butovsky O, Gilbert JA, Sisodia SS. Sex-specific effects of microbiome perturbations on cerebral Aβ amyloidosis and microglia phenotypes. J Exp Med. 2019 Jul 1;216(7):1542-1560. Epub 2019 May 16 PubMed.
Kundu P, Torres ER, Stagaman K, Kasschau K, Okhovat M, Holden S, Ward S, Nevonen KA, Davis BA, Saito T, Saido TC, Carbone L, Sharpton TJ, Raber J. Integrated analysis of behavioral, epigenetic, and gut microbiome analyses in AppNL-G-F, AppNL-F, and wild type mice. Sci Rep. 2021 Feb 25;11(1):4678. PubMed.
Magnusson KR, Hauck L, Jeffrey BM, Elias V, Humphrey A, Nath R, Perrone A, Bermudez LE. Relationships between diet-related changes in the gut microbiome and cognitive flexibility. Neuroscience. 2015 May 14;300:128-140. PubMed.
Raber J, Fuentes Anaya A, Torres ER, Lee J, Boutros S, Grygoryev D, Hammer A, Kasschau KD, Sharpton TJ, Turker MS, Kronenberg A. Effects of Six Sequential Charged Particle Beams on Behavioral and Cognitive Performance in B6D2F1 Female and Male Mice. Front Physiol. 2020;11:959. Epub 2020 Aug 28 PubMed.
Torres ER, Akinyeke T, Stagaman K, Duvoisin RM, Meshul CK, Sharpton TJ, Raber J. Effects of Sub-Chronic MPTP Exposure on Behavioral and Cognitive Performance and the Microbiome of Wild-Type and mGlu8 Knockout Female and Male Mice. Front Behav Neurosci. 2018;12:140. Epub 2018 Jul 18 PubMed.
Leviatan S, Segal E. Identifying gut microbes that affect human health. Nature. 2020 Nov;587(7834):373-374. PubMed.
Limeta A, Ji B, Levin M, Gatto F, Nielsen J. Meta-analysis of the gut microbiota in predicting response to cancer immunotherapy in metastatic melanoma. JCI Insight. 2020 Dec 3;5(23) PubMed.
Gut Microbiome Manipulation May Facilitate Immunotherapy Response. Cancer Discov. 2021 Feb;11(2):221. Epub 2020 Dec 18 PubMed.
Make a Comment
To make a comment you must login or register.