What Makes a Microglia? Tales from the Transcriptome
Quick Links
Microglia have come under close scrutiny as their role in neurodegenerative disease becomes ever more apparent; alas, studying these finicky cells from people has not been easy. In the May 25 Science, researchers led by Christopher Glass, University of California, San Diego, release the first-ever comprehensive analysis of the human microglial transcriptome. They find it largely matches that from mice. They also report that the signature quickly fades when human microglia are taken from the brain and placed in culture. The same is true for mouse microglia, as described in a May 17 Neuron paper from Ben Barres and colleagues at Stanford University School of Medicine, California.
While the papers bring little solace for researchers trying to culture microglia, Barres and colleagues define factors that will at least keep the cells alive in the absence of serum, which contains factors foreign to the microglial environment. They also report that injecting cultured mouse microglia back into mouse brains restores their normal gene expression, suggesting these highly dynamic cells do not change irreversibly when taken out of their realm. Together, the two papers will help scientists create better microglial models, researchers agreed.
“Glass’ is the first deep characterization of microglia directly from people,” said Oleg Butovsky, Brigham and Women’s Hospital, Boston, who was not involved in either study. It makes sense that microglia are extremely plastic in response to the environment, as these cells are the sensors of the brain, he said. “The study will be useful as a source for those labs working to manipulate microglia in an effort to change their phenotype and work out potential therapeutic or diagnostic approaches.”
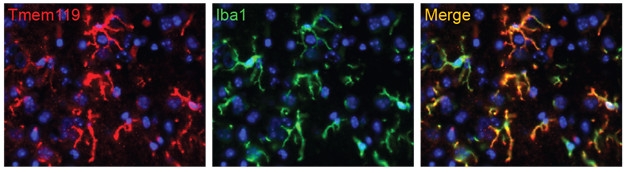
Home Sweet Home: Microglia (green) that previously lost their identity in culture express microglial genes such as Tmem119 (red) when injected back into a mouse brain. [Neuron, Bohlen et al.]
Recently, scientists have turned to human induced pluripotent stem cells (iPSCs) to try to derive microglia that better approximate those in the brain (see Jul 2016 news; Abud et al., 2017). However, they have no epigenetic or transcriptional blueprint to compare them against, said David Gosselin, co-first author on the Glass paper. To better define human microglia, he and fellow co-first authors Dylan Skola and Nicole Coufal took donated tissue from 19 people undergoing brain surgery to treat epilepsy, a brain tumor, or stroke. For each patient, the researchers took the healthiest tissue sample and extracted the cells within a few hours.
Next, the authors sequenced the mRNA to determine which genes were expressed. They found a microglial signature comprising 881 transcripts that were expressed at least 10-fold higher than in whole cortical brain tissue. Hundreds of those same genes are up- or downregulated in diseases such as Alzheimer’s, frontotemporal lobar disorder, and schizophrenia. Interestingly, microglia highly expressed genes, including TREM2, SORL1, MEF2C, INPP5D, and CD33, which have been associated with AD.
The signature was similar among samples taken from different parts of the same brain sections, from samples taken from the same patient a year apart, and among the 19 subjects, regardless of the disease they had. The human transcriptome also largely overlapped with that from 10-week-old mouse microglia. More than 13,000 of the almost 16,000 orthologous gene pairs were expressed at similar levels. However, the human microglia highly expressed 400 genes that mouse microglia did not, while the latter expressed 263 genes to a greater extent.
The two species seem to similarly regulate microglial transcription according to two techniques that identify transcriptionally active DNA. Assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq), which pinpoints the accessible regions of chromatin, and chromatin immunoprecipitation (ChIP)-sequencing, which identifies active transcription factor binding sites, yielded similar patterns for mouse and human microglia. Open and active areas of DNA correlated with enhancer regions for the microglia-specific genes. The transcription factor recognition motifs within those DNA regions were also similar. The transcriptional mechanisms are largely conserved between species, the authors wrote.
How does that microglial signature change in vitro? The Glass group previously reported that mouse microglia kept in culture for seven days drastically reduced expression of hundreds of microglia-specific genes (see Gosselin et al., 2014). To find out if the same happened in human microglia, the authors compared samples taken directly from the brain to those kept in culture for seven days. They looked strikingly different, with the cultured cells upregulating 300 genes more than 10-fold, including those for inflammation and stress responses, and downregulating more than 700, including those for immune cell function and brain development. Almost 300 of those 1,000 dysregulated genes have been associated with various neurodegenerative diseases. These differences arose as early as six hours after culture, and lasted the entire seven days.
Gosselin tried but could not find tissue culture conditions that preserved the microglial gene expression pattern. Even microglia from induced pluripotent stem cells, which are supposed to resemble in vivo cells better than their isolated counterparts, expressed a pattern more similar to the in vitro microglia than those from the brain. It is likely that three-dimensional cultures that include several brain cell types will be necessary to get closer to the in vivo gene profile, the authors wrote.
“For the first time, we are providing a unique perspective and a good understanding of what makes microglia a microglia from a genomic and epigenomic perspective,” Gosselin told Alzforum. “Our efforts are a good validation of the mouse microglia models, and will probably be helpful in better understanding their limitations and strengths.”
Monica Carson, University of California, Riverside, agreed. “If you want to make in vitro systems more like in vivo, you now have the molecular and transcriptional targets you know you can manipulate,” she said. This will help guide researchers in making better cultured models, she said. “Now we know what we need them to be like.” She added that it will be important in the future to sample across ethnicities, brain regions, and development. Butovsky added that he would be interested to know how the microglial transcriptome changes in various disease states. For instance, how do cells surrounding Aβ plaques in AD compare to those near lesions in multiple sclerosis?
“It’s an incredibly robust and systematic data set,” wrote Josh Morganti, University of Kentucky, Lexington, to Alzforum. It reinforces the validity of using mouse models to define translatable mechanisms of microglia function, and suggests that in vitro experiments involving primary microglia are futile at the moment, given the loss of microglia identity, he said.
The paper from the Barres lab backs up the idea that taking microglia out of the mouse brain and culturing them changes their gene expression almost immediately. An hour after cells had been cultured, first author Christopher Bohlen and colleagues saw that microglia started to change the expression of 1,300 genes, turning off those associated with mature microglia in the brain and instead expressing those typical of immature microglia. Many of those upregulated genes are also expressed highly in diseases such as AD and amyotrophic lateral sclerosis, said Chris Bennett, second author on the paper.
The researchers also reported that microglia exposed to serum divided quickly and became more phagocytic. Serum is typically added to a growth medium to keep cells alive, but contains a lot of factors foreign to the natural microglial environment. To try to culture these cells serum-free, the researchers started with conditioned media used to grow astrocytes, which can keep microglia alive in co-culture. They found three factors that kept microglia alive, including TGF-β2, CSF-1, and cholesterol. However, even cells treated with these factors failed to express a normal microglial signature. “We need to be extremely mindful of conclusions we draw from cultured microglia,” said Bennett. The research group plans to seek other factors that would get the cells closer to an in vivo profile in culture.
“This work may completely change the way we have been culturing primary microglial cells in vitro,” said Shanya Jiang, University of New Mexico, Albuquerque. She noted that based on Barres’ data, gene transcription may already have changed in the ex vivo microglia Glass and colleagues isolated, since they were exposed to serum for several hours.
“It is clear that microglia acquire different features as soon as they are put in culture and that they retain only minimal transcriptome and epigenetic features found in the original cells,” wrote Marco Colonna, Washington University in St. Louis, to Alzforum.
Interestingly, if Bohlen took cultured microglia that had reverted to an immature state and injected them back into a mouse brain, the cells began to look more like their old selves again, expressing genes such as TMEM119 (see image above). “That suggests that microglia have a great deal of plasticity,” said Bennett. “If we know that microglia can undergo these dramatic changes that resemble disease, but then can return to normal, that’s quite encouraging for the development of microglia-specific therapies,” he added.—Gwyneth Dickey Zakaib
References
News Citations
Paper Citations
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, Caraway CA, Fote GM, Madany AM, Agrawal A, Kayed R, Gylys KH, Cahalan MD, Cummings BJ, Antel JP, Mortazavi A, Carson MJ, Poon WW, Blurton-Jones M. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017 Apr 19;94(2):278-293.e9. PubMed.
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014 Dec 4;159(6):1327-40. PubMed.
Further Reading
Primary Papers
- Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JC, Sajti E, Jaeger BN, O'Connor C, Fitzpatrick C, Pasillas MP, Pena M, Adair A, Gonda DD, Levy ML, Ransohoff RM, Gage FH, Glass CK. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017 Jun 23;356(6344) Epub 2017 May 25 PubMed.
- Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB, Barres BA. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron. 2017 May 17;94(4):759-773.e8. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
University of Kentucky
This is an incredibly robust and systematic dataset from Chris Glass’ group. A few salient points regarding the manuscript stand out to me. 1.) The conservation of a transcriptional repertoire between mouse and human microglia is a key finding. Importantly, this reinforces the validity of using mouse models of brain disorders to define translatable mechanisms related to microglia function in the etiology and/or progression of those disorders. 2.) Comparison of the human microgliome with whole-tissue analyses showed that microglia significantly express multiple risk alleles associated with several progressive neurodegenerative disorders. These findings bolster the putative link between resident innate immune response in the diseased brain. 3.) The use of in vitro experiments involving primary microglia is a frustratingly futile endeavor given the loss of microglia identity and acquisition of a quasi-phenotype (similar to previous work by Butovsky et al., 2014). This is incredibly important in terms of microglia-specific therapeutic development for neurodegenerative disorders, and is unfortunate as the penultimate testbed would be to generate human iPSC-derived true microglia harboring specific risk alleles for AD, PD, MS, etc.…More
One of the main caveats with this paper is the use of a “catch-all” microglia FACS enrichment strategy. The authors even acknowledge this caveat in a sense with their dataset’s inability to show variations in transcripts across the multiple diseases/diagnosis/age. Additionally, cells were only taken from cortical tissue, and recent work shows that there is a regional heterogeneity of microglia gene expression (Grabert et al., 2016).
Two important questions remain in my mind: How does this gene signature change between different subsets of microglia within a diseased brain? Do the enriched motifs/genes in microglia have a functional basis in disease, as some of them do in homeostatic function (e.g., Bialas and Stevens, 2013)?
References:
Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci. 2014 Jan;17(1):131-43. Epub 2013 Dec 8 PubMed.
Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM, McColl BW. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 2016 Mar;19(3):504-16. Epub 2016 Jan 18 PubMed.
Bialas AR, Stevens B. TGF-β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat Neurosci. 2013 Dec;16(12):1773-82. Epub 2013 Oct 27 PubMed.
Make a Comment
To make a comment you must login or register.