Scanning Top 20 GWAS Hits for Amyloidosis: Slim Pickings
Quick Links
To identify genes responsible for amyloid deposition, researchers led by Liana Apostolova of Indiana University in Indianapolis looked for correlations between variants of the top 20 AD risk genes and brain amyloid in 977 adults. In the January 16 JAMA Neurology, they report that after ApoE4, a variant in the lipid transporter ABCA7 most tightly associated with brain amyloid. Links to other gene variants depended on disease stage, with asymptomatic amyloid carriers, people with mild cognitive impairment, and those with dementia showing various degrees of correlation. That this analysis failed to pull out other genes previously linked to plaques, such as CR1, highlights the complexity of amyloidosis, suggested researchers. “There are many ways to cause amyloid to accumulate, so I am not surprised that Apostolova and colleagues saw what they did,” noted Sam Gandy, Mount Sinai Medical Center, New York.
- Scientists analyzed variants in 20 AD risk genes relative to brain amyloidosis.
- After ApoE4, the strongest link was with ABCA7.
- FERMT2 correlated with amyloid only in people with MCI.
Apostolova focused on 27 single nucleotide polymorphisms (SNPs) scattered among 20 genes that had been identified previously in AD GWAS, or linked to brain amyloidosis (Oct 2013 news). Prior studies had looked at one or two variants at a time, and in small numbers of people, whereas Apostolova used multivariate regression models to study the contributions of all the variants at once. The 977 volunteers had had a florbetapir PET scan and genotyping as part of the Alzheimer Disease Neuroimaging Initiative (ADNI). The group averaged 74 years old and had nearly as many men as women.
After controlling for age, sex, education, the strong effect of ApoE4, and the smaller effects of all other variants, ABCA7 and two other genes still significantly associated with whole-brain florbetapir uptake. They are EPHA1, which encodes a tyrosine kinase receptor, and PICALM, the gene for a vesicle-assembly protein that regulates production and clearance of Aβ and tau (Xiao et al., 2012; Moreau et al., 2014). Both EPHA1 and PICALM have been linked to amyloid before (May 2015 news; Hughes et al., 2014).
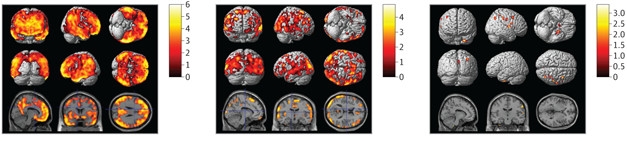
Local Associations: Brighter colors show where SNPs in ABCA7 (left), EPHA1 (center), and PICALM (right) influence amyloid deposition. [From Apostolova et al., ©2018 American Medical Association. All rights reserved.]
The large sample size gave Apostolova an opportunity to search for associations in specific stages of disease. Among the volunteers, 322 were cognitively normal, 496 had mild cognitive impairment, and 159 dementia. When the investigators reran the regression models on each group separately, up came a variant in the fermitin family homologue 2 (FERMT2) gene as having an effect only in people with MCI. ABCA7’s effects were stronger in the early stages, both normal and MCI, as well. In dementia, new associations emerged between florbetapir binding and variants in SORL1, ZCWPW1, DSG2, and CLU. In the MCI subgroup, the SORL1 variants also correlated with PET signals.
To Apostolova’s knowledge, this is the first time FERMT2 has been implicated in amyloid deposition. Scientists know little about the gene. It encodes a β3-integrin coactivator expressed in the brain, and may play a role in inflammation and leucocyte trafficking. A homolog suppresses tau toxicity in Drosophila (Shulman et al., 2013).

Moving Target: The association of FERMT2 SNP, rs17125944, with florbetapir binding varies among normal (left), MCI (middle), and AD (right) groups. Red indicates the highest positive correlation coefficients; blue increasingly negative correlations. [From Apostolova et al., ©2018 American Medical Association. All rights reserved.]
In an unbiased, genome-wide screen for siRNAs that modulate amyloid precursor protein metabolism in APP-overexpressing cells, the group of Philippe Amouyel, Institut Pasteur, Lille, France, identified a silencer of FERMT2 as the top hit (Chapuis et al., 2017). Subsequently, the French researchers found SNPs in FERMT2, including rs17125944, associated with cerebrospinal fluid Aβ levels in 2,886 people with AD. “What we have observed in metabolism also seems to apply in the clinic to this biomarker,” Amouyel told Alzforum.
Apostolova examined the relationship between genetic variation and where amyloid is in the brain. A voxel-based study associated ABCA7 with sparsely distributed voxel clusters in the frontal gyri and left parietal lobe in normal controls; there were many more voxels throughout the brain in people with MCI, but none at all in in the dementia group. In the MCI group, FERMT2 associated with a large cluster of voxels centered on the left middle temporal gyrus.
The results strengthen the idea that ABCA7 variants primarily affect early amyloidosis, while other variants contribute to different aspects of AD pathology, Apostolova said. Previously, when her group associated variants with brain metabolism or gray-matter atrophy, ABCA7 was not among the top hits, and FERMT2 was absent (Stage et al., 2016). “FERMT2 did not pop up in our previous analysis, and we had larger sample sizes there, so it does not seem to be relevant to neurodegeneration per se,” Apostolova said. Other groups, too, have tied distinct genes to amyloid versus neurodegeneration (Sep 2016 conference news).
The study provides the largest and most comprehensive look at how genetic variants affect amyloid deposition, but even so it is limited by its size, noted John Hardy, University College, London. “We now have GWAS hits for Alzheimer’s disease based on huge numbers of samples, more than 75,000 cases and controls. It is natural we should want to test these SNPs against other phenotypes that may be directly related to the AD phenotype, or indeed be related to entirely different outcomes,” Hardy wrote in an email to Alzforum. However, he noted that the number of samples studied with such phenotypes remains very small, which limits statistical power. “I would regard this as an exploratory study, which sets an agenda for study when we have larger samples,” he wrote.—Pat McCaffrey
References
News Citations
- Paper Alert: New Alzheimer’s Genes Published
- New Role For PICALM: Flushing Aβ From the Brain
- Amyloid and Neurodegeneration Have Different Underlying Genetics
Paper Citations
- Xiao Q, Gil SC, Yan P, Wang Y, Han S, Gonzales E, Perez R, Cirrito JR, Lee JM. Role of Phosphatidylinositol Clathrin Assembly Lymphoid-Myeloid Leukemia (PICALM) in Intracellular Amyloid Precursor Protein (APP) Processing and Amyloid Plaque Pathogenesis. J Biol Chem. 2012 Jun 15;287(25):21279-89. PubMed.
- Moreau K, Fleming A, Imarisio S, Lopez Ramirez A, Mercer JL, Jimenez-Sanchez M, Bento CF, Puri C, Zavodszky E, Siddiqi F, Lavau CP, Betton M, O'Kane CJ, Wechsler DS, Rubinsztein DC. PICALM modulates autophagy activity and tau accumulation. Nat Commun. 2014 Sep 22;5:4998. PubMed.
- Hughes TM, Lopez OL, Evans RW, Kamboh MI, Williamson JD, Klunk WE, Mathis CA, Price JC, Cohen AD, Snitz BE, Dekosky ST, Kuller LH. Markers of cholesterol transport are associated with amyloid deposition in the brain. Neurobiol Aging. 2014 Apr;35(4):802-7. Epub 2013 Oct 3 PubMed.
- Shulman JM, Imboywa S, Giagtzoglou N, Powers MP, Hu Y, Devenport D, Chipendo P, Chibnik LB, Diamond A, Perrimon N, Brown NH, De Jager PL, Feany MB. Functional screening in Drosophila identifies Alzheimer's disease susceptibility genes and implicates Tau-mediated mechanisms. Hum Mol Genet. 2013 Oct 9; PubMed.
- Chapuis J, Flaig A, Grenier-Boley B, Eysert F, Pottiez V, Deloison G, Vandeputte A, Ayral AM, Mendes T, Desai S, Goate AM, Kauwe JS, Leroux F, Herledan A, Demiautte F, Bauer C, Checler F, Petersen RC, Blennow K, Zetterberg H, Minthon L, Van Deerlin VM, Lee VM, Shaw LM, Trojanowski JQ, Albert M, Moghekar A, O'Brien R, Peskind ER, Malmanche N, Schellenberg GD, Dourlen P, Song OR, Cruchaga C, Amouyel P, Deprez B, Brodin P, Lambert JC, ADGC, Alzheimer’s Disease Neuroimaging Initiative. Genome-wide, high-content siRNA screening identifies the Alzheimer's genetic risk factor FERMT2 as a major modulator of APP metabolism. Acta Neuropathol. 2017 Jun;133(6):955-966. Epub 2016 Dec 8 PubMed.
- Stage E, Duran T, Risacher SL, Goukasian N, Do TM, West JD, Wilhalme H, Nho K, Phillips M, Elashoff D, Saykin AJ, Apostolova LG. The effect of the top 20 Alzheimer disease risk genes on gray-matter density and FDG PET brain metabolism. Alzheimers Dement (Amst). 2016;5:53-66. Epub 2016 Dec 19 PubMed.
Further Reading
Primary Papers
- Apostolova LG, Risacher SL, Duran T, Stage EC, Goukasian N, West JD, Do TM, Grotts J, Wilhalme H, Nho K, Phillips M, Elashoff D, Saykin AJ, Alzheimer’s Disease Neuroimaging Initiative. Associations of the Top 20 Alzheimer Disease Risk Variants With Brain Amyloidosis. JAMA Neurol. 2018 Mar 1;75(3):328-341. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.