RNA-DNA Pairs: At the Root of C9ORF72 Repeat Damage?
Quick Links
Researchers have uncovered a new explanation for the chaos caused by a hexanucleotide repeat expansion in the C9ORF72 gene, the most common genetic cause of sporadic and familial amyotrophic lateral sclerosis and frontotemporal dementia. In the March 6 issue of Nature, Jiou Wang and scientists at Johns Hopkins University in Baltimore report that during transcription, the sense strand of the six-lettered repeat twists up on itself, leaving the opposite strand free. The antisense strand then binds the nascent C9ORF72 mRNA and the RNA-DNA hybrid disrupts normal transcription. “We propose a molecular cascade that leads from abnormalities in the DNA structures to patient pathology,” Wang told Alzforum. “We believe that these DNA structures and RNA-DNA duplexes are the root cause for both loss of the full-length C9ORF72 products, as well as the build-up of toxic RNAs.”
Previous reports suggest that the GGGGCC repeat expansion in C9ORF72 causes abnormal transcription, translation, and accumulation of C9ORF72 RNA, as well as partial loss of the protein. Similar guanine-rich regions have been known to form structures called G-quadruplexes in both DNA and RNA (see Bochman et al., 2012). In DNA, these form when a single strand folds back on itself to make a four-sided structure held together by hydrogen bonds between the guanines (see image below). Similar structures are found in G-rich RNA, and are suspected of promoting aggregation of C9ORF72 mRNA (see Fratta et al., 2012; Nov 2013 news story). Wang’s group wondered if the structures form in the C9ORF72 repeat DNA as well.

G-Quadruplexes.
DNA and RNA sequences can form stable structures (at right in the image) stabilized by bonds among guanines at the four corners of a quadruplex. [Images courtesy of Haeusler et al., 2014. Nature.]
To find out, first author Aaron Haeusler and colleagues probed the repeat using several biochemical and biophysical techniques, including one called dimethyl sulfate footprinting, which can pick up a signature of G-quadruplexes through DNA sequencing. All tests revealed evidence of the structures in the C9ORF72 gene.
To determine whether the quadruplexes have any function, the authors created a simple plasmid-based expression system in a test tube and examined how varying numbers of repeats affected mRNA transcription. The more repeats a plasmid had, the more it produced stunted mRNA instead of full-length product.
It took some detective work to figure out if the G-quadruplexes halted transcription, but the researchers had a hunch that the structures allow RNA-DNA binding. When strands of a DNA helix separate during processes such as transcription, RNA may bind, but usually transiently. However, if a G-quadruplex twists up one strand, it stabilizes the opening, and leaves the other strand free for more stable RNA binding. The researchers thought that might derail C9ORF72 transcription. Sure enough, when they added an enzyme that digests RNA-DNA hybrids to the in-vitro expression system, the number of full-length transcripts rose while the incomplete ones petered out.
To check whether the same process could occur in human cells, the authors looked at extracts from patients’ motor cortexes and spinal cords. Hauesler found that people with ALS who carried the C9ORF72 expansion had truncated transcripts. These incomplete fragments of RNA contained repeats that give rise to G-quadruplexes and RNA hairpins. Since proteins are known to bind to such secondary structures, the authors wondered if the truncated RNAs could be toxic.
Which proteins might the RNAs bind? An RNA pull-down assay isolated 288 candidates. One was nucleolin, a protein that helps synthesize and assemble ribosomes inside the nucleolus, a small sphere in the nucleus dedicated to this purpose. In cells from patients with a C9ORF72 repeat expansion, however, nucleolin spread throughout the nucleus (see image below). These patient-derived cells also succumbed more easily to stress. Interestingly, if the researchers introduced truncated C9ORF72 RNAs into normal human embryonic kidney cells, nucleolin spread across the nucleus and the cells died prematurely.
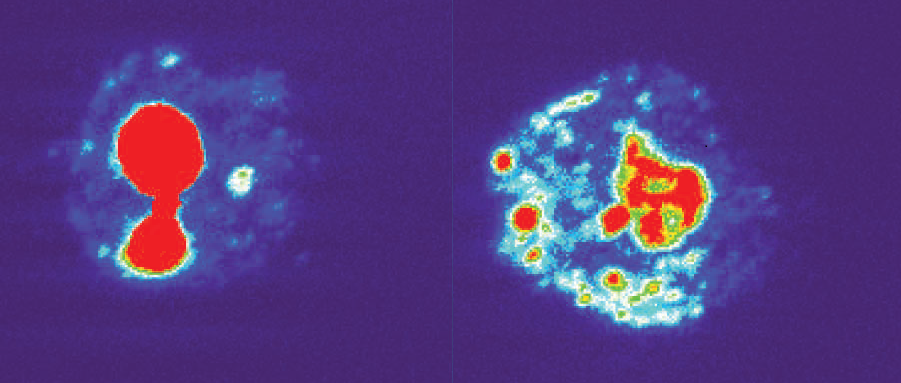
Runaway Nucleolin. Rather than remain inside the nucleolus as it normally does (left), nucleolin strays out into the nucleus in cells with the C9ORF72 repeat (right). Image courtesy of Haeusler et al., 2014. Nature.
Based on these findings, Wang and colleagues propose a model in which G-quadruplexes in C9ORF72 repeat regions lead to RNA-DNA pairs, which halt mRNA transcription early. The resulting stunted mRNAs sequester crucial RNA-binding proteins and lead to cell stress (see schematic below). This could explain several observations about C9ORF72-related damage, said Wang. The incomplete transcription reduces C9ORF72 protein, while the truncated transcripts contribute to RNA aggregates. He speculated that the shortened transcripts are the ones that yield small repeat peptides that were recently found in mutation carriers (see Feb 2013 news story).

The big picture. Quadruplexes formed by the C9ORF72 DNA (grey) cause RNA (red) transcription to abort. RNA quadruplexes then sequester nucleolar proteins (NCL) and also yield toxic poly-dipeptides (purple), when translated in the cytoplasm. Image courtesy of Haeusler et al., 2014. Nature.
“This [proposed mechanism] suggests a double whammy,” said Christopher Pearson, Hospital for Sick Children, Toronto. “Not only is there seemingly a loss of function because of incomplete transcription, but since the aborted G-quadruplex-containing transcript is toxic, there’s a gain of function as well.” Scientists have long debated whether the C9ORF72 mutation results in a loss or gain of function. While the authors did not show that the RNA-DNA hybrids form in vivo, Pearson says enough evidence exists for these structures in trinucleotide repeat-expansion diseases (see Reddy et al., 2011) that it is not hard to believe they crop up in the C9ORF72 expansions. Wang said the results suggest that if a small molecule could break apart the RNA-DNA hybrids, it could lessen toxic effects. He plans to study this possibility.
“A highlight is that nucleolin interacts with RNA G-quadruplexes formed by C9ORF72,” wrote J. Paul Taylor, St. Jude Children’s Research Hospital, Memphis, Tennessee, in an accompanying News and Views article. Adrian Isaacs, University College London, agreed. “This identifies nucleolar stress as a potentially novel pathway for neurodegeneration in C9ORF72-related FTD and ALS and possibly other repeat-expansion diseases,” he wrote to Alzforum via email. He wonders, however, whether the stress results directly from the repeats or if it is a more general marker of stress in neurodegeneration.
The findings bear striking similarities to a recent study by scientists including Samie Jaffrey, Cornell University, New York, who found that the CGG repeat expansion in the Fragile X gene forms RNA-DNA duplexes that block transcription (see Feb 2014 news story). Jaffrey and colleagues traced that gene silencing to DNA methylation. Scientists previously observed hypermethylation in the C9ORF72 promoter (see Jun 2013 news story). Whether RNA-DNA hybrids lead to those epigenetic changes remains to be seen, said Wang.
“These papers are pointing us in a new direction for understanding this large set of neurological diseases,” said Jaffrey. “A common feature may be the formation of these RNA-DNA duplexes that then activate cellular signaling pathways that ultimately control the expression of the repeat-associated genes,” he explained. He would be interested to know how the C9ORF72 repeats affect its epigenetics.—Gwyneth Dickey Zakaib
References
News Citations
- Sense, Antisense: C9ORF72 Makes Both Forms of RNA, Peptides
- RNA Twist: C9ORF72 Intron Expansion Makes Aggregating Protein
- Gene Silencing in Fragile X Syndrome Explained?
- Methylation a Turn Off for Disease Gene C9ORF72?
Paper Citations
- Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012 Nov;13(11):770-80. Epub 2012 Oct 3 PubMed.
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. PubMed.
- Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, Nichol Edamura K, Wang YH, Pearson CE. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011 Mar;39(5):1749-62. Epub 2010 Nov 4 PubMed.
Further Reading
Papers
- Belzil VV, Bauer PO, Gendron TF, Murray ME, Dickson D, Petrucelli L. Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res. 2014 Feb 12; PubMed.
- Zamiri B, Reddy K, Macgregor RB Jr, Pearson CE. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J Biol Chem. 2014 Feb 21;289(8):4653-9. Epub 2013 Dec 26 PubMed.
- Stepto A, Gallo JM, Shaw CE, Hirth F. Modelling C9ORF72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis and frontotemporal dementia. Acta Neuropathol. 2014 Mar;127(3):377-89. Epub 2013 Dec 24 PubMed.
Primary Papers
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014 Mar 13;507(7491):195-200. Epub 2014 Mar 5 PubMed.
- Taylor JP. Neurodegenerative diseases: G-quadruplex poses quadruple threat. Nature. 2014 Mar 13;507(7491):175-7. Epub 2014 Mar 5 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.