Missing Ingredient: New Mice Model Alzheimer’s Genetic Variability
Quick Links
A person’s genetic background promotes Alzheimer’s or protects against it, but existing mouse models in genetically identical strains ignore this aspect of a multigenic disease in humans, an outbred population. Is this why mouse treatment studies have failed to translate and the field has been backing away from mouse models? To address these questions head-on, researchers developed a new kind of AD model, where multiple strains of genetically diverse mice mimic the diversity of people.
- New Alzheimer’s mice model the genetic diversity found in people.
- Easily produced for drug testing, lines may better translate to LOAD.
- Work draws road map for richer modeling of polygenic disease.
In a study spearheaded by Catherine Kaczorowski of the Jackson Laboratory in Bar Harbor, Maine, investigators crossed the 5XFAD mouse with 27 genetically diverse strains. This created a panel of mice that all express the same causative mutations in APP and presenilin genes, but in the context of different protective or deleterious gene variants elsewhere in the genome. Comparison with existing 5XFAD mice suggests the new strains, as a group, better reflect the variation in both the pathology and cognitive changes of human LOAD. This may improve the translation of research from mouse to human going forward, Kaczorowski believes. Her work appeared December 27 in Neuron.
To generate the new lines, first author Sarah Neuner started with the widely used 5XFAD transgenic model, which carries human APP and PS1 with multiple autosomal-dominant AD mutations. She mated heterozygous 5XFAD females with males from 27 different BXD genetic reference strains. All of the BXDs are themselves products of crosses between C57BL/6J (B6) and DBA/2J (D2)—two common strains that differ at just over 5 million sites in their genomes. A product of more than 45 years' work, the strains offer a rich and open resource for genetic studies in mice, with complete genetic information and extensive phenotyping available online at GeneNetwork.org. Half of the offspring of the 5XFAD/BXD cross carried the dominant 5XFAD transgene, and half did not, and so the researchers created both AD and their isogenic control mice with one cross.
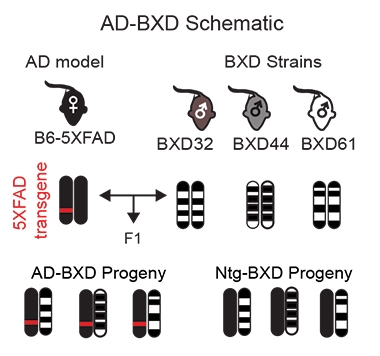
Variety Show. Mating 5XFAD mice with BXD strains produced offspring with AD genes on a varied genetic background. Half the progeny carried no FAD genes, and served as non-transgenic controls. [From Neuner et al., 2018 Neuron.]
The investigators measured memory, motor function and anxiety in all the mice, and tracked brain Aβ1-42 accumulation. As expected, across all strains, 5XFAD carriers developed memory deficits at an earlier age than their non-transgenic littermates, and their deficits in contextual fear tests were greater at older ages. However, the effect of the transgene varied from strain to strain—in some, 5XFAD accelerated memory decline; in others, it did little. This is similar to how genetic variation affects age of onset in people with FAD mutations (Ryman et al., 2014), the authors note. Strains also varied in the extent of cortical Aβ1-42 and plaque deposition, although, as in people, amyloid accumulation and cognitive decline did not correlate.
The results were not explained by strain differences in transgene expression, or expression of mouse APP or presenilin genes, which all held steady across the strains and were unaffected by age or sex. What did account for the strain differences? For one, ApoE genotype and sex were linked to poorer cognitive function in the 5XFAD mice, similar to what’s seen in people (Altmann et al., 2014; Mielke et al., 2014). But more interestingly, other genes modified the disease phenotype. In people, a genetic risk score, aka polygenic risk score or PRS, that takes into account the cumulative small effects of many risk genes can predict risk of late-onset AD (Mar 2017 news). For the mice, the investigators contrived a similar score, based on 21 genes known to confer risk of AD in people. For each mouse strain, its risk score correlated with memory performance at 14 months, but only in transgenic mice. The risk score did not track with amyloid deposition, sensorimotor skills or anxiety. This suggests the mouse gene variants, similar to their human counterparts, specifically modified the risk of cognitive decline in mice that already have AD pathology, i.e., they influenced a mouse’s ability to withstand a head full of amyloid.

Polygenic Risk. In transgenic (left panel), but not in non-transgenic mice (right panel), a genetic risk score (GRS) based on human AD risk genes predicts declining memory in 14-month-old mice of different strains indicated by number. [Courtesy of Neuner et al., 2018, Neuron.]
The investigators also documented variations in expression of specific AD risk genes across strains. Transcriptome-wide changes in the strains followed a similar, and familiar, pattern: expression of genes associated with neuronal activity, structure and function went down, while immune response genes were boosted. The consistent upregulation of immune pathway genes known to occur in humans and in the 5XFAD strains did not occur in non-transgenic mice, suggesting it is a feature of AD and not of normal aging in these mice.
The results uncovered an unrecognized resilience in the original, and popular, 5XFAD line, which is maintained in B6 mice. The mouse has moderately high levels of Aβ42, but has a low genetic risk score and maintains cognitive function better than other strains. Kaczorowski told Alzforum her lab is actively working to identify the resilience factors at work in this strain. The result illustrates how evaluating just this one strain, or any one strain, in preclinical studies would hamper translation to humans. When evaluating drugs, testing multiple strains, including ones with a higher genetic risk scores and robust cognitive decline, may offer better translation to humans, particularly for late-onset AD, she said.
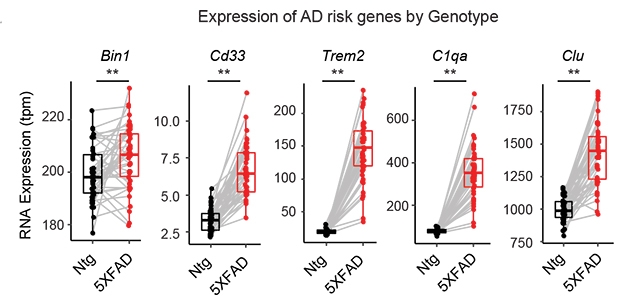
Diversity Rules. Expression of other AD risk genes varies greatly among 5XFAD carrier strains, demonstrating the effect of genetic background. [Courtesy of Neuner et al., 2018, Neuron.]
The models are a resource for the AD research community, Kaczorowski told Alzforum. Individual phenotyping data is publicly available on the Accelerating Medicines Partnership (AMP-AD) Knowledge Portal and other public databases. On the GeneNetwork site, the data sit side by side with deep phenotyping and complete genetic information for the BXD strains, along with analysis tools that are open to anyone “It’s important to disseminate the data early and often, and to encourage people to reach out with ideas for collaboration,” she said.
Her group currently is characterizing a total of 70 AD-BXD strains, and will release all the behavioral, transcriptional, and pathology data as it is generated. They also have stored frozen brain tissue and peripheral organs from the mice, which are available for study. The mice themselves are sold through the Jackson Laboratory.—Pat McCaffrey
References
Research Models Citations
News Citations
Paper Citations
- Ryman DC, Acosta-Baena N, Aisen PS, Bird T, Danek A, Fox NC, Goate A, Frommelt P, Ghetti B, Langbaum JB, Lopera F, Martins R, Masters CL, Mayeux RP, McDade E, Moreno S, Reiman EM, Ringman JM, Salloway S, Schofield PR, Sperling R, Tariot PN, Xiong C, Morris JC, Bateman RJ, Dominantly Inherited Alzheimer Network. Symptom onset in autosomal dominant Alzheimer disease: a systematic review and meta-analysis. Neurology. 2014 Jul 15;83(3):253-60. Epub 2014 Jun 13 PubMed.
- Altmann A, Tian L, Henderson VW, Greicius MD, Alzheimer's Disease Neuroimaging Initiative Investigators. Sex modifies the APOE-related risk of developing Alzheimer disease. Ann Neurol. 2014 Apr;75(4):563-73. Epub 2014 Apr 14 PubMed.
- Mielke MM, Vemuri P, Rocca WA. Clinical epidemiology of Alzheimer's disease: assessing sex and gender differences. Clin Epidemiol. 2014;6:37-48. Epub 2014 Jan 8 PubMed.
Other Citations
External Citations
Further Reading
No Available Further Reading
Primary Papers
- Neuner SM, Heuer SE, Huentelman MJ, O'Connell KM, Kaczorowski CC. Harnessing Genetic Complexity to Enhance Translatability of Alzheimer's Disease Mouse Models: A Path toward Precision Medicine. Neuron. 2019 Feb 6;101(3):399-411.e5. Epub 2018 Dec 27 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
UTHSC
In general, teams who are interested in robust results—Catherine Kaczorowski’s and many others—study larger numbers of strains/genomes, with fewer replicates within strain. If a team can afford to study 400 cases, then studying four each of 100 genotypes is about the sweet spot—perhaps two males and two females of each strain. One could bump that up to eight each of 50 genotypes if interested in two time points.
Imagine a human geneticist with the luxury of studying four identical twins (and in male and female versions). At some point, the replication within strain/genome buys you less and less information. Replication in the range from two to eight lets you detect potential problems, such as outliers and errors. But the point is not to understand any one strain. The point is to generate robust data that will generalize (for more, see Williams and Williams, 2017; Williams, 2009). …More
We are making this work easier for teams: 1. they do not need to do any genotyping; 2. they have easy access to deep electronic health records of all of these genotypes of mice; 3. they have access to the whole statistical workflow at a single website (gn2.genenetwork.org).
The typical functional genomics study (i.e. asking "what does this gene do?") will use 10 to 20 experimental knockout/transgenic animals on one genetic background per treatment group. Perhaps as many controls. This provides results that have good statistical power but not good generality and translatability. However, some of the 5XFAD x BXD lines will be cool to use in more focused studies—particularly interventions that delay or prevent age-related decline in performance.
References:
Williams RW, Williams EG. Resources for Systems Genetics. Methods Mol Biol. 2017;1488:3-29. PubMed.
Williams RW. Herding cats: the sociology of data integration. Front Neurosci. 2009;3(2):154-6. Epub 2009 Sep 15 PubMed.
Make a Comment
To make a comment you must login or register.