Islands of Mutated Neurons Dot the Brain. Are They Bad for Us?
Quick Links
The genomes we inherit from our parents are not set in stone. After sperm meets egg, and mitosis is on its merry way, mutations occur in our DNA. They give rise to populations of cells harboring unique genomes and indeed, researchers suspect that cliques of brain cells bearing variants could trigger neurodegeneration. But how common are such somatic mutations? According to a study in the October 15 Nature Communications, everyone’s brain is chock-full of them.
- Deep sequencing of 102 genes in postmortem brain uncovered 39 unique somatic variants.
- Those variants were detected in half of the brains sampled.
- Algorithm predicts that pockets of pathogenically mutated neurons exist in every person’s brain.
Researchers led by Patrick Chinnery at the University of Cambridge, U.K., deep-sequenced genes related to neurodegenerative disorders from several regions of 54 postmortem brains from people with Alzheimer’s, Parkinson’s disease, or controls. They found 39 unique somatic variants lurking in those samples, which collectively represent a minuscule fraction of the human brain. Extrapolating their findings to the entire brain, the scientists predicted that people have between 100,000 and 1 million cells carrying somatic mutations in genes related to neurodegenerative disease. These variants are spread out into hundreds of small islands and occasional large ones. The number of brains in the study was too small to definitively tie such mutations to neurodegenerative disease, but the predictions imply a potential role.
“In a very thorough and carefully planned study, Chinnery’s team have significantly advanced our knowledge of brain somatic mutations, and possible relevance to neurodegenerative diseases,” wrote Christos Proukakis of University College London. “They demonstrate that ‘islands’ of neurons with somatic mutations may occur frequently in the human brain, and larger studies will be needed to show if there is a clear relationship with neurodegenerative disease.”

Islands in the Brain. The distribution of somatic variants across the 54 brains. Large circles represent the number of sequenced DNA molecules in each region; smaller circles reflect the occurrence of a given single (green) or multiple (purple) region variant. [Courtesy of Keogh et al., Nature Communications, 2018.]
Researchers have long suspected that the brain contains a genomic patchwork of cells harboring mutations that arose at different stages of development. These variants have even been tied to a handful of sporadic cases of neurodegenerative disease (Beck et al., 2004; Jul 2015 news; Nicolas et al., 2018).
However, due to the localized nature of these mutations throughout the brain, tracking them down, let alone investigating their involvement in disease, requires cutting-edge sequencing, cell isolation, and computational techniques. Using single-cell sequencing, a recent study estimated that each cell in the brain harbors 200–400 somatic mutations that arose during brain development, while another study reported around 1,500 per post-mitotic neurons (Bae et al., 2018; Lodato et al., 2015). The mutation rate of human neurons also reportedly ramps up with age (Lodato et al., 2018). Yet the cumulative impact of these mutations, and how many cells harbor each one, remains uncertain.
To address these questions, first author Michael Keogh and colleagues employed ultra-deep sequencing of select genes in different regions from postmortem brain samples. The scientists resequenced each sample more than 1,000 times, allowing them to detect variants with high specificity and sensitivity, even for genes that are typically extremely difficult to sequence. Then, using a computational model of brain development, they used their findings to estimate the burden of somatic variation in the entire brain. The researchers sequenced the exons of 56 genes linked to neurodegenerative disease, including APP, PS1/2, TREM2, ApoE, LRRK2, and Parkin, as well as 46 cancer-related genes expressed at low levels in the brain as controls. They sequenced these genes in DNA extracted from 173 frozen brain samples, which came from the frontal cortices, entorhinal cortices, cingulate cortices, cerebella, or medullas of 54 brains. Twenty of the brains came from donors with AD, 20 from people with Lewy body disorders, and 14 from age-matched controls. The researchers extracted the bulk DNA from a section of each region, so could not discern if a variant they found came from neurons, glia, or vascular cells. In addition, they sequenced blood samples that were available from six of the donors.
In all, the researchers found 39 somatic variants among 44 of the 173 brain samples (see image above). Eight were in neurodegenerative disease genes: EIF4G1, LRRK2, NOTCH3, SETX, SORL1, TAF15, UCHL1, and VPS35. Twenty-seven of the 54 brains in the cohort had at least one variant. Of the 39, 18 appeared only in a single region of one brain, and four were detected in a single blood sample. These single-region mutations (SRMs) were similarly common in any of the tested brain regions, in neurodegenerative disease versus control genes, or in people with or without neurodegenerative disease. That’s not to say these variants are not linked to neurodegeneration. The authors estimated they would need to analyze 190 cases and 190 controls to detect a twofold difference in variant incidence between groups.
Seventeen of the 39 variants occurred in more than one brain region, or in paired blood/brain samples. Unlike the SRMs, these multiple-region mutations (MRMs) overwhelmingly were in cancer-related genes. Only one MRM was detected in a neurodegenerative disease-related gene—TAF15—and it was detected in three brain regions of one healthy control. Fifteen of the remaining 16 MRMs resided within genes related to myeloproliferative blood disorders, and blood samples had a higher frequency of the mutant alleles than did brain samples from the same donor (see image below). This suggested circulating immune cells were the source of the somatic mutant alleles.
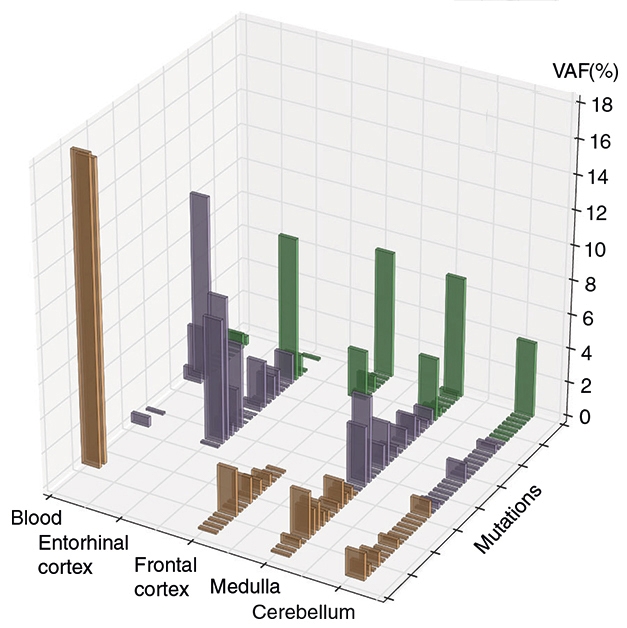
Somatic Variation. Among people with AD (brown bars), Lewy body disease (purple), and controls (green), each of 39 mutations occurred at different rates across four brain regions and blood. [Courtesy of Keogh et al., 2018.]
The two most common MRMs occurred in the genes for TET2 and DNMT3A, which play a role in DNA methylation and have been implicated in hematopoietic disorders. In a blood sample from one AD patient, two TET2 variants cropped up in almost 20 percent of sequences (see image above). Forty percent of brains from people with Lewy body disorders had one of these mutations, compared with just 7 percent of brains from healthy controls. The researchers speculated that this could explain previously reported observations that people with PD have altered DNA methylation in the blood and brain. Circulating cells carrying these variants might infiltrate the brain and wreak havoc, they proposed. In support of this, the occurrence of these TET2 and DNMT3A variants was highest in brain regions harboring Lewy body pathology.
The researchers next sought to extrapolate their findings to estimate the burden of variants in neurodegenerative disease-related genes across the entire brain. Using a cellular barcoding technique, they estimated they had sequenced DNA from around 611,000 cells. They were also able to estimate the proportion of cells in any given region that carried a somatic mutation in a neurodegenerative disease-related gene.
They fed this data into a statistical algorithm that simulated brain development to predict the total number and distribution of mutated cells among the estimated 86 billion in each brain. The answer: 100,000 to 1 million cells carry a somatic mutation in a disease-related gene. Incorporating information about how cells divide, differentiate, and mutate during development, the algorithm also foretold that each person likely had one large island of 10,000 to 100,000 cells that grew from one original mutation in a disease gene, while 10 percent of people had at least one island of more than 200,000 such cells. In addition, each brain contained 75 to 481 smaller islands, each consisting of just more than 100 descendants of a cell carrying a pathological variant.
The researchers speculated that these islands of somatic variants trigger sporadic neurodegenerative disease, which reportedly affects roughly 10 percent of the human population.
“Even if only one island of 10,000 to 100,000 cells carried a mutation related to Alzheimer's disease, it could hypothetically serve as a hotspot for amyloid or tau seed formation and could in turn have an effect that spreads beyond their own cell population,” commented Huntington Potter of the University of Colorado in Denver. However, he noted that the data do not yet conclusively link such mutations and disease. In AD patients, the authors found no variants in the entorhinal cortex, a region affected early in the disease process.
While Chinnery and colleagues speculated that larger numbers of mutated cells would be more likely to cause disease, they did not exclude the possibility that a small number would be sufficient to instigate the slow spread throughout the brain of toxic protein, such as Aβ, α-synuclein, tau, and others that can propagate by templated misfolding.
Gaël Nicolas of Radboud University Medical Center in Nijmegen, The Netherlands, called the study a significant contribution to the field of somatic mutations in the brain, but cautioned that it fell short of linking specific somatic variants to aberrant protein aggregation or neurodegeneration, as did several other recent reports, including his own.
Alexander Hoischen, also at Radboud, had similar impressions. “The study shows how the latest sequencing technology combined with smart analysis can detect somatic mutations much more accurately,” he wrote. “This paper provides further leads about fascinating human biology, but elucidating the exact involvement of somatic mutations in brain disease, and neurodegeneration in particular, requires a lot of work.”—Jessica Shugart
References
News Citations
Paper Citations
- Beck JA, Poulter M, Campbell TA, Uphill JB, Adamson G, Geddes JF, Revesz T, Davis MB, Wood NW, Collinge J, Tabrizi SJ. Somatic and germline mosaicism in sporadic early-onset Alzheimer's disease. Hum Mol Genet. 2004 Jun 15;13(12):1219-24. Epub 2004 Apr 28 PubMed.
- Nicolas G, Acuña-Hidalgo R, Keogh MJ, Quenez O, Steehouwer M, Lelieveld S, Rousseau S, Richard AC, Oud MS, Marguet F, Laquerrière A, Morris CM, Attems J, Smith C, Ansorge O, Al Sarraj S, Frebourg T, Campion D, Hannequin D, Wallon D, Gilissen C, Chinnery PF, Veltman JA, Hoischen A. Somatic variants in autosomal dominant genes are a rare cause of sporadic Alzheimer's disease. Alzheimers Dement. 2018 Dec;14(12):1632-1639. Epub 2018 Aug 13 PubMed.
- Bae T, Tomasini L, Mariani J, Zhou B, Roychowdhury T, Franjic D, Pletikos M, Pattni R, Chen BJ, Venturini E, Riley-Gillis B, Sestan N, Urban AE, Abyzov A, Vaccarino FM. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2018 Feb 2;359(6375):550-555. Epub 2017 Dec 7 PubMed.
- Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D'Gama AM, Cai X, Luquette LJ, Lee E, Park PJ, Walsh CA. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015 Oct 2;350(6256):94-98. PubMed.
- Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, Sherman MA, Vitzthum CM, Luquette LJ, Yandava CN, Yang P, Chittenden TW, Hatem NE, Ryu SC, Woodworth MB, Park PJ, Walsh CA. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018 Feb 2;359(6375):555-559. Epub 2017 Dec 7 PubMed.
Further Reading
Papers
- Leija-Salazar M, Piette C, Proukakis C. Review: Somatic mutations in neurodegeneration. Neuropathol Appl Neurobiol. 2018 Apr;44(3):267-285. Epub 2018 Feb 28 PubMed.
- Lanoiselée HM, Nicolas G, Wallon D, Rovelet-Lecrux A, Lacour M, Rousseau S, Richard AC, Pasquier F, Rollin-Sillaire A, Martinaud O, Quillard-Muraine M, de la Sayette V, Boutoleau-Bretonniere C, Etcharry-Bouyx F, Chauviré V, Sarazin M, le Ber I, Epelbaum S, Jonveaux T, Rouaud O, Ceccaldi M, Félician O, Godefroy O, Formaglio M, Croisile B, Auriacombe S, Chamard L, Vincent JL, Sauvée M, Marelli-Tosi C, Gabelle A, Ozsancak C, Pariente J, Paquet C, Hannequin D, Campion D, collaborators of the CNR-MAJ project. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017 Mar;14(3):e1002270. Epub 2017 Mar 28 PubMed.
Primary Papers
- Keogh MJ, Wei W, Aryaman J, Walker L, van den Ameele J, Coxhead J, Wilson I, Bashton M, Beck J, West J, Chen R, Haudenschild C, Bartha G, Luo S, Morris CM, Jones NS, Attems J, Chinnery PF. High prevalence of focal and multi-focal somatic genetic variants in the human brain. Nat Commun. 2018 Oct 15;9(1):4257. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
UCL
In a very thorough and carefully planned study, Patrick Chinnery’s team from Cambridge has significantly advanced our knowledge of brain somatic mutations, and their possible relevance to neurodegenerative diseases.
The strengths of the study include using two different strategies for capture and very high coverage targeted sequencing. Results were validated with the second method, which uses unique molecular identifiers to help discount any PCR and other errors. They used several brain regions from two very common neurodegenerative disorders (Alzheimer’s and Lewy body disorders). Once the somatic frequency was known, the authors conducted very elegant mathematical modeling of brain development, taking into account key parameters such as programmed developmental cell death, and asymmetric cell divisions. They demonstrate that “islands” of neurons with somatic mutations may occur frequently in the human brain, and larger studies will be needed to show if there is a clear relationship with neurodegenerative disease.…More
This is not a single-cell study, so it avoids the possible caveats of whole-genome amplification, although it does not allow inference of the cell type(s) which had a mutation. Using paired blood samples where available, and including genes involved in myeloproliferative disorders, shows a hematopoietic origin for mutations in some genes, even when they were also found in DNA extracted from brain.
This may well be just the tip of the iceberg, as only one type of mutation (single nucleotide) is detectable by this approach. Other studies have shown somatic copy-number variations in the brain, including associations with genes relevant to Alzheimer’s and Parkinson’s diseases. An important point to always remember is that we cannot know if neurons that died due to a person’s neurodegenerative illness had additional mutations. There are exciting times ahead.
References:
Mokretar K, Pease D, Taanman JW, Soenmez A, Ejaz A, Lashley T, Ling H, Gentleman S, Houlden H, Holton JL, Schapira AH, Nacheva E, Proukakis C. Somatic copy number gains of α-synuclein (SNCA) in Parkinson's disease and multiple system atrophy brains. Brain. 2018 Jun 15; PubMed.
Bushman DM, Kaeser GE, Siddoway B, Westra JW, Rivera RR, Rehen SK, Yung YC, Chun J. Genomic mosaicism with increased amyloid precursor protein (APP) gene copy number in single neurons from sporadic Alzheimer's disease brains. Elife. 2015 Feb 4;4 PubMed.
Rouen University Hospital
With this very nice report, Keogh et al. contribute significantly to the developing field of somatic mutations in the brain. There has been huge progress during the last few years on this topic, thanks to the advent of deep massive parallel sequencing, the development of single-molecule capture techniques, and the improvement of bioinformatics detection algorithms. Here, the authors report somatic mutations in several brain samples, which probably arose during fetal development. They sequenced different bulk brain regions in several individuals, not distinguishing whether mutations occurred in neurons or glial cells. This reports adds to the recent data showing 200–400 somatic variants per cerebral cell during brain development based on single-cell sequencing and fetal samples (Bae et al., 2018) and an unsuspected high yield of ~1,500 somatic variants per post-mitotic neuron (Lodato et al., 2015). …More
With the hypothesis that some of these mutations may trigger neurodegenerative diseases by producing abnormal proteins that would spread throughout the brain, several other groups, including ours, have attempted to detect somatic variants which could be putatively interpreted as pathogenic (Sala Frigerio et al., 2015; Nicolas et al., 2018). Although we identified a few SORL1 somatic variants that might have contributed to AD development, no clearly pathogenic variant was identified in the autosomal dominant AD-causative genes in these studies, including this new study by Keogh et al.
Here, somatic mutations occurring in unique regions (single regional mutations), of likely developmental origin, did not occur more frequently in neurodegenerative genes than in cancer genes, nor did they affect more cases than controls, although the authors acknowledge that this part missed statistical power for a formal conclusion. So far, efforts to identify putatively neurodegenerative disease-triggering somatic variants have remained unsuccessful, despite the recent big steps in technology and knowledge. The authors also identified multiple regional mutations, which were enriched in hematopoiesis genes, suggesting clonal hematopoiesis of myeloid precursors and blood-brain barrier dysfunction. They hypothesize that some of them could have contributed to the development of neurodegenerative diseases. This remains hypothetical and will require both validation with increased numbers of brain samples of patients and controls, and further investigation.
Overall, this report adds significant knowledge on the natural history of somatic mutations in the brain, including mutational rates, and opens new doors for the development of this research field in the context of neurodegenerative diseases.
References:
Bae T, Tomasini L, Mariani J, Zhou B, Roychowdhury T, Franjic D, Pletikos M, Pattni R, Chen BJ, Venturini E, Riley-Gillis B, Sestan N, Urban AE, Abyzov A, Vaccarino FM. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2018 Feb 2;359(6375):550-555. Epub 2017 Dec 7 PubMed.
Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D'Gama AM, Cai X, Luquette LJ, Lee E, Park PJ, Walsh CA. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015 Oct 2;350(6256):94-98. PubMed.
Sala Frigerio C, Lau P, Troakes C, Deramecourt V, Gele P, Van Loo P, Voet T, De Strooper B. On the identification of low allele frequency mosaic mutations in the brains of Alzheimer's disease patients. Alzheimers Dement. 2015 Apr 29; PubMed.
Nicolas G, Acuña-Hidalgo R, Keogh MJ, Quenez O, Steehouwer M, Lelieveld S, Rousseau S, Richard AC, Oud MS, Marguet F, Laquerrière A, Morris CM, Attems J, Smith C, Ansorge O, Al Sarraj S, Frebourg T, Campion D, Hannequin D, Wallon D, Gilissen C, Chinnery PF, Veltman JA, Hoischen A. Somatic variants in autosomal dominant genes are a rare cause of sporadic Alzheimer's disease. Alzheimers Dement. 2018 Dec;14(12):1632-1639. Epub 2018 Aug 13 PubMed.
Radboud University Medical Center
The study by Keogh et al. shows how the latest sequencing technology combined with smart analysis can detect somatic mutations much more accurately. With this, the authors deliver extra evidence that, contrary to dogma from old textbooks, “DNA is not the same in every cell of the human body.” This was previously also shown for single neurons, different brain regions, and several other tissues of the human body. This paper provides further leads about fascinating human biology, but the exact involvement of somatic mutations in brain disease, and in neurodegeneration in particular, requires a lot of future work.
University of Colorado Alzheimer’s and Cognition Center
Every Brain is a Genetic Mosaic: Implications for Neurodegeneration
Cell division, including the key steps of DNA replication and chromosome segregation, must be extremely precise in order to assure the accurate, complete, and equal replication and transfer of genetic information from one generation to another (meiosis), and from one cell to its daughter cells during development (mitosis). Defects in this process can lead to many disorders, including cancer, developmental disorders, and possibly neurodegenerative diseases. Such defects fall into two major categories: 1) mutations, including single nucleotide changes and segmental duplications, inversions, translocations, and deletions, and 2) chromosome mis-segregation that results in aneuploidy. Both can lead to cancer, but the latter is perhaps best illustrated by Down's syndrome/trisomy 21, the most common developmental disorder that arises from chromosome mis-segregation.…More
Genetic change/damage at the level of single nucleotides or of DNA segments or at the level of whole chromosomes can occur both during meiosis, leading to the whole organism being affected, and during mitosis, leading to mosaic aneuploidy. Substantial work using fluorescence in situ hybridization (FISH) and other techniques has shown that aneuploidy (i.e., an abnormal number of chromosomes) in neurons is commonly associated with neurological disorders, including both familial and sporadic Alzheimer’s disease, frontotemporal dementia, Niemann-Pick Type C, and autism (Potter, 1991; Geller and Potter, 1999; Rossi et al., 2013; Yurov et al., 2007; Granic and Potter, 2013; Boeras et al., 2018; Arendt et al., 2010; Caneus et al., 2017; Potter et al., 2016; Migliore et al., 1999; Mosch et al., 2007). Such aneuploidy likely underlies the majority of cell death in Alzheimer’s disease and in at least one form of frontotemporal dementia (Arendt et al., 2010; Caneus et al., 2017). Furthermore, different levels and types of aneuploidy may contribute to the clinical heterogeneity associated with many different neurodegenerative disorders and observed in people with Down’s syndrome (Potter, 2016).
Because DNA sequencing, and especially single-cell DNA sequencing, involves multiple steps that can introduce errors, such as single nucleotide mutations, it has been extremely difficult to assess the overall rate and accumulation of low levels of somatic mutations during development and aging. Several laboratories have addressed these difficulties, for example, by improving the fidelity of the amplification and sequencing steps, and by extending the depth of sequencing in order to reduce false-positive rates, and by assessing not only single cells, but also progeny of dividing stem cells (Bae et al., 2018; Werner and Sottoriva, 2018; Huang et al., 2018; Verjeijen et al., 2018; Lodato et al., 2018). In this paper, Keogh and colleagues very elegantly focused on identifying somatic mutations in specific genes that may contribute to human disease. Specifically, they sequenced a total of 102 genes, including 46 genes associated with cancer and 56 genes associated with neurodegenerative disorders at >5,000-fold depth in 173 adult human brain regions from 20 Alzheimer’s disease brains, 20 Lewy body disease brains, and 14 brains without significant neuropathology, some of which could be paired with blood samples, thus allowing any observed somatic mutations to be validated. Using various mathematical models, they were able to calculate the mutation rate and could thus predict that islands of a few hundred mutated neurons are likely to be quite common in the general population. Even if only one such island of ~10,000–100,000 cells, which they predict should happen once in each individual, carried a mutation related to Alzheimer's disease, such islands could hypothetically serve as hotspots for amyloid or tau seed formation and could in turn have an effect that spreads beyond their own cell population. Curiously, the levels of mosaic single-region mutations did not differ between the three populations, which somewhat reduces the likelihood that these mutations may underlie a large number of sporadic neurodegenerative diseases. Nonetheless, the appreciation that the brain is a mosaic patchwork of clonal islands of cells whose genotypes have changed during development and/or during aging is an important insight for disease diagnosis and therapy, and also potentially for understanding brain aging more generally.
References:
Potter H. Review and hypothesis: Alzheimer disease and Down syndrome--chromosome 21 nondisjunction may underlie both disorders. Am J Hum Genet. 1991 Jun;48(6):1192-200. PubMed.
Geller LN, Potter H. Chromosome missegregation and trisomy 21 mosaicism in Alzheimer's disease. Neurobiol Dis. 1999 Jun;6(3):167-79. PubMed.
Rossi G, Conconi D, Panzeri E, Redaelli S, Piccoli E, Paoletta L, Dalprà L, Tagliavini F. Mutations in MAPT gene cause chromosome instability and introduce copy number variations widely in the genome. J Alzheimers Dis. 2013;33(4):969-82. PubMed.
Yurov YB, Vorsanova SG, Iourov IY, Demidova IA, Beresheva AK, Kravetz VS, Monakhov VV, Kolotii AD, Voinova-Ulas VY, Gorbachevskaya NL. Unexplained autism is frequently associated with low-level mosaic aneuploidy. J Med Genet. 2007 Aug;44(8):521-5. Epub 2007 May 4 PubMed.
Granic A, Potter H. Mitotic spindle defects and chromosome mis-segregation induced by LDL/cholesterol-implications for Niemann-Pick C1, Alzheimer's disease, and atherosclerosis. PLoS One. 2013;8(4):e60718. PubMed.
Boeras DI, Granic A, Padmanabhan J, Crespo NC, Rojiani AM, Potter H. Alzheimer's presenilin 1 causes chromosome missegregation and aneuploidy. Neurobiol Aging. 2008 Mar;29(3):319-28. PubMed.
Arendt T, Brückner MK, Mosch B, Lösche A. Selective cell death of hyperploid neurons in Alzheimer's disease. Am J Pathol. 2010 Jul;177(1):15-20. PubMed.
Caneus J, Granic A, Chial HJ, Potter H. Using Fluorescence In Situ Hybridization (FISH) Analysis to Measure Chromosome Instability and Mosaic Aneuploidy in Neurodegenerative Diseases. In: Frade JM, Gage FH, editors. Genomic Mosaicism in Neurons and Other Cell Types, 2017
Potter H, Granic A, Caneus J. Role of Trisomy 21 Mosaicism in Sporadic and Familial Alzheimer's Disease. Curr Alzheimer Res. 2016;13(1):7-17. PubMed.
Migliore L, Botto N, Scarpato R, Petrozzi L, Cipriani G, Bonuccelli U. Preferential occurrence of chromosome 21 malsegregation in peripheral blood lymphocytes of Alzheimer disease patients. Cytogenet Cell Genet. 1999;87(1-2):41-6. PubMed.
Mosch B, Morawski M, Mittag A, Lenz D, Tarnok A, Arendt T. Aneuploidy and DNA replication in the normal human brain and Alzheimer's disease. J Neurosci. 2007 Jun 27;27(26):6859-67. PubMed.
Potter H. Beyond Trisomy 21: Phenotypic Variability in People with Down Syndrome Explained by Further Chromosome Mis-segregation and Mosaic Aneuploidy. J Down Syndr Chromosom Abnorm. 2016;2(1) Epub 2016 Mar 31 PubMed.
Bae T, Tomasini L, Mariani J, Zhou B, Roychowdhury T, Franjic D, Pletikos M, Pattni R, Chen BJ, Venturini E, Riley-Gillis B, Sestan N, Urban AE, Abyzov A, Vaccarino FM. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2018 Feb 2;359(6375):550-555. Epub 2017 Dec 7 PubMed.
Werner B, Sottoriva A. Variation of mutational burden in healthy human tissues suggests non-random strand segregation and allows measuring somatic mutation rates. PLoS Comput Biol. 2018 Jun;14(6):e1006233. Epub 2018 Jun 7 PubMed.
Huang AY, Yang X, Wang S, Zheng X, Wu Q, Ye AY, Wei L. Distinctive types of postzygotic single-nucleotide mosaicisms in healthy individuals revealed by genome-wide profiling of multiple organs. PLoS Genet. 2018 May;14(5):e1007395. Epub 2018 May 15 PubMed.
Verheijen BM, Vermulst M, van Leeuwen FW. Somatic mutations in neurons during aging and neurodegeneration. Acta Neuropathol. 2018 Jun;135(6):811-826. Epub 2018 Apr 28 PubMed. Correction.
Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, Sherman MA, Vitzthum CM, Luquette LJ, Yandava CN, Yang P, Chittenden TW, Hatem NE, Ryu SC, Woodworth MB, Park PJ, Walsh CA. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018 Feb 2;359(6375):555-559. Epub 2017 Dec 7 PubMed.
Make a Comment
To make a comment you must login or register.