GPS for the Brain—New Publicly Available Atlas Pinpoints Gene Expression
Quick Links
Google™ may help you find a street address, and Google Earth might help you view it, but if you are looking to map gene expression in the brain, then you need something with finer resolution. BGEM, or Brain Gene Expression Map, can help you ogle that gene. Curator Tom Curran and colleagues at St. Jude Children’s Research Hospital, Memphis, Tennessee, have opened BGEM to the public. The freely available resource should be a boon to researchers studying the brain.
Curran and colleagues explain all in a March 28 paper in PLoS Biology. First author Susan Magdaleno and colleagues report that the database has over 30,000 images covering thousands of mouse genes. For each gene there are four expression maps taken at embryonic days 11.5 and 15.5, at postnatal day 7, and from adult brain. Because all the images are taken from radioactive in-situ hybridizations, the atlas provides greater sensitivity for genes expressed at low levels, and it has better signal-to-noise than other atlases compiled from non-radioactive hybridization images. The BGEM database identifies brain expression patterns for genes that have previously been mapped to only non-neuronal tissues, so there is new information in there that can be mined.
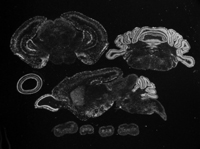
BGEM Leaves You Reelin
A representative image from the Brain Gene Expression Map database shows expression of the protein reelin in day 7 postnatal brain tissue from the mouse. The radioactive probes give high contrast, good for detecting those weakly expressed genes. This image is just one of over 30,000 freely available ones from the BGEM website. [Image courtesy of Tom Curran, manager of BGEM at St. Jude Children’s Research Hospital.]
The atlas is currently being used to select candidate genes for the GENSAT (Gene Expression Nervous System Atlas) project, which aims to document expression patterns of all the genes in the nervous system. But BGEM, and its images, can now be used by anyone provided he or she cites BGEM and the PLoS paper (see below). In fact, researchers at St. Jude Children’s Research Hospital have already used the database to link developmental gene expression to tumors arising from specific anatomical locations in the brain. This has led to the hypothesis that radial glia cells act as stem cells for some ependymomas (see Taylor et al., 2005).
BGEM is fully searchable and has up-to-date information from NCBI. The database is linked to PubMed, OMIM (Online Mendelian Inheritance in Man), GENSAT, and other databases. In fact, BGEM was designed to be used in conjunction with them. “Users can view gene expression patterns in BGEM and compare them with high-resolution images from GENSAT, and they can request BAC reporter mice from the Mutant Mouse Regional Resource Centers directly through the linked Web sites,” write Magdaleno and colleagues. Users can also combine BGEM information with data from other studies, such as gene chip analysis. “The BGEM database could be emulated by others to make large datasets conveniently interoperable. We look forward to linking our image sets to other gene expression databases that are actively growing,” write the authors.—Tom Fagan
References
Paper Citations
- Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P, Magdaleno S, Dalton J, Calabrese C, Board J, Macdonald T, Rutka J, Guha A, Gajjar A, Curran T, Gilbertson RJ. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005 Oct;8(4):323-35. PubMed.
External Citations
Further Reading
No Available Further Reading
Primary Papers
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, Norland SM, Rice DS, Dosooye N, Shakya S, Mehta P, Curran T. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006 Apr;4(4):e86. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.