Genomic Double-Stranded RNA: Does C9ORF72 Cause Viral Mimetic Disease?
Quick Links
A telltale sign of viral invasion, double-stranded RNA sounds cellular alarm bells that touch off a firestorm of interferon. According to a paper published July 7 in Science Translational Medicine, dsRNAs that arise from non-viral sources, including hexanucleotide expansions in the C9ORF72 gene, can also instigate harmful inflammation. Researchers led by Mark Albers at Massachusetts General Hospital in Boston spotted dsRNAs mingling with TDP-43 inclusions in the cytoplasm of neurons in C9ORF72 mutation carriers who had died with amyotrophic lateral sclerosis and frontotemporal dementia.
- Cytoplasmic double-stranded RNA detected in neurons and glia of C9ORF72 mutation carriers with ALS/FTD.
- cdsRNA co-localized with TDP-43 inclusions.
- In human cells and mice, cdsRNA unleashed a deadly interferon cascade
- cdsRNA traveled between connected neurons, propagating death.
In cultured human cells, and in a mouse model burdened with cytoplasmic dsRNAs, the RNA couplets provoked a dramatic interferon response, akin to that triggered by a virus. The resulting inflammation killed not only neurons harboring the dsRNAs, but also connected neurons. What’s more, the researchers reported that the dsRNAs traveled between connected neurons. FDA-approved JAK inhibitors stanched the IFN response and spared neurons.
According to the study, cytoplasmic dsRNA (cdsRNA) may propagate neurodegeneration in people with C9-ALS/FTD. Albers hypothesizes that the findings could extend to other forms of neurodegenerative disease marked by TDP-43 dysfunction, including other types of ALS and FTD, and many cases of AD. This is because this RNA-binding protein is tasked with keeping complementary dsRNA-prone sequences from pairing up.
Brian Freibaum, St. Jude’s Children’s Hospital, Memphis, Tennessee, agreed that the findings cast cytoplasmic dsRNA as a likely contributor to the ill effects of C9ORF72 hexanucleotide expansions, and possibly other forms of neurodegenerative disease as well.
First, some background on this unorthodox biology. Hexanucleotide expansions in the first intron of the C9ORF72 gene are transcribed from both sense and antisense strands, as well as in multiple reading frames, leading to five possible transcripts. These either aggregate into RNA foci or, via a process called repeat-associated non-ATG (RAN) translation, get translated into dipeptide repeats, which themselves aggregate. In a recently reported feed-forward loop, the repeat transcripts instigated a cellular stress response that, in turn, promoted RAN translation (Nov 2018 conference news). Because both sense and antisense strands are transcribed, Albers and colleagues wondered if those transcripts might be subject to yet a third fate: Could complementary strands pair up into dsRNA?
First author Steven Rodriguez and colleagues addressed this question by hunting for dsRNA in postmortem brain samples from eight C9ORF72 mutation carriers with FTD/ALS, and six controls. Using an antibody specific for dsRNA, the researchers detected elevated amounts in the cytoplasm of neurons, microglia, and astrocytes in the frontal cortices, motor cortices, and cerebella of C9 patients but not controls.
Separately, cytoplasmic inclusions of TDP-43, an RNA-binding protein that carries out many of its duties in the nucleus, are a pathological hallmark of C9-ALS/FTD. Strikingly, the scientists found that cdsRNA co-localized with cytoplasmic TDP-43 inclusions. When Rodriguez sequenced the dsRNAs, more than half encoded C9 repeat sequences.

Cytoplasmic Liaisons. In the frontal (upper row) and motor cortices (bottom row) of a person with C9-ALS/FTD, dsRNA (green) mingled with phospho-TDP-43 (red) in the cytoplasm of different cell types. Sytox (blue) labels the nucleus. [Courtesy of Rodriguez et al., Science Translational Medicine, 2021.]
Albers told Alzforum that ongoing experiments aim to uncover the identity of the other dsRNAs. He hypothesizes that TDP-43 dysfunction could lead to the formation of cdsRNA from repeat and inversion-laden sequences, including C9ORF72 repeats. Specifically, his group is hunting for cdsRNA in other forms of neurodegenerative disease marked by TDP-43 pathology, including sporadic ALS/FTD and AD. Notably, the researchers did not detect substantial cdsRNA in two ALS patients with SOD1 mutations, a form of this disease without TDP-43 inclusions.
In keeping with the presence of cdsRNA in the C9-ALS/FTD samples, the researchers detected evidence of a robust Type 1 IFN response, including elevated phospho-PKR, a known interferon-stimulated gene. Genes involved in dsRNA-binding and Type 1 IFN signaling were among the most upregulated in previously published RNA sequencing data from the cerebella of people with C9-ALS/FTD.
When transfected into the cytoplasm, dsRNA encoding the C9ORF72 repeats also proved deadly to cultured human cells. Within two days of transfection, human ReNcell VM neuroprogenitors—a cell type that expresses markers of neurons, astrocytes, and oligodendrocytes—ramped up expression of interferon-stimulated proteins, including MDA5, PKR, and phospho-STAT1. Then they started to die.
Treatment of the cells with FDA-approved JAK inhibitors ruxolitinib, baricitinib, or tofacitinib blocked the phosphorylation of STAT1 and spared neurons. Approved to treat various inflammatory disorders including rheumatoid arthritis, these drugs are known to counter Type 1 IFN signaling. Baricitinib has FDA emergency use authorization to treat COVID-19, as well, and tofacitinib lowered the risk of respiratory failure or death in people hospitalized with COVID-19 in a recent trial (Guimarães et al., 2021).
To further investigate the neurotoxic effects of cytoplasmic dsRNA, the scientists used mice. They took advantage of two transgenic models, Nd1 and Nd2, in which the inserted human APP transgene had inadvertently undergone a slew of genomic acrobatics, including inversions. Curiously, these mice displayed profound neurodegeneration, while mice expressing the same transgene without the structural snafus did not. Type 1 interferon signaling was also rampant in their brains, hence Rodriguez et al. suspected dsRNA shenanigans.
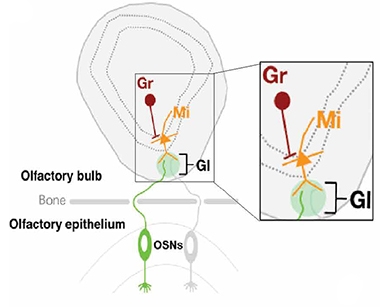
Olfactory Anatomy. Axons project from olfactory sensory neurons (OSNs) in the nose to the olfactory bulb in the brain, where they synapse with mitral cells (Mi) in the glomerulus (Gl). The Mi connect to granule neurons (Gr). Only OSNs express the transgene that leads to cdsRNA formation in Mi and Gr. [Courtesy of Rodriguez et al., Science Translational Medicine, 2021.]
In a previous study, they had generated Nd1 and Nd2 mice that conditionally expressed the transgene in but 1 percent olfactory sensory neurons, peripheral neurons in the nose that connect to neurons in the olfactory bulb within the brain. In the current study, they confirmed that indeed, cdsRNA was expressed in the olfactory epithelium in mice expressing the inverted transgenes there.
The scientists used these mice to ask how cdsRNAs within a small fraction of neurons would influence IFN signaling and neurodegeneration in the brain. In a nutshell, they detected a whopping interferon response in both the peripheral olfactory sensory neurons that expressed the wacky transgene, as well as in the connected olfactory bulb. Numbers of microglia also doubled in the OB. The researchers detected cdsRNA produced from the transgene within axon terminals of the OB, as well as within OB neurons. This suggested that the dsRNA had traveled down OSN axons and crossed into connected cells.
Strikingly, although so few OSNs expressed the transgene and resulting cdsRNA, the transgenic mice had 40 percent fewer OSNs and 20 percent fewer OB neurons than did controls by 3 months of age. This dramatic cell death was rescued when the mice were treated with the JAK inhibitor ruxolitinib.
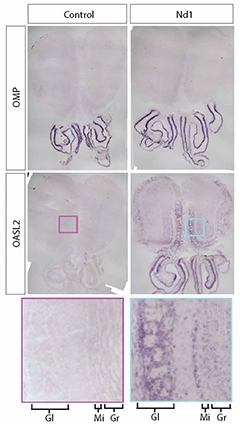
Invading Interferon. Olfactory sensory neurons, marked by OMP staining (top) in both controls (left) and Nd1 mice (right). In Nd1 mice, only OSNs express the APP transgene that produces cdsRNA. The interferon-stimulated gene Oasl2 is expressed in OSNs and in the connected olfactory bulb in Nd1 mice. Enlarged excerpt shows layers of olfactory bulb (Gl, glomerulus; Mi, mitral cells; Gr, granule cells). [Courtesy of Rodriguez et al., Science Translational Medicine, 2021.]
Overall, the mouse data suggested that cdsRNA produced by even a tiny fraction of neurons is capable of wreaking havoc in the brain. The researchers are in the process of defining the relative contributions to this deadly cascade of cdsRNA propagation, IFN signaling, and microglia. That ruxolitinib spared neurons suggests that the IFN response deals the deadly blow.
The mouse data implicate cdsRNA as a general inflammatory instigator; indeed, Albers believes that any scenario that promotes cdsRNA formation could prove harmful to neurons. In the case of C9-ALS/FTD, dsRNA produced from the repeat sequences contributes to a pool of dsRNA, while malfunctions in RNA-binding proteins such as TDP-43 may exacerbate its accumulation. However, dsRNA can also arise from other sources, including the de-repression of transposable elements, Albers noted. TDP-43 keeps these ancient genomic relics under wraps, so theoretically any disorder marked by TDP-43 dysfunction could lead to more cdsRNAs (Saldi et al., 2014; Tam et al., 2019).
Benjamin Wolozin of Boston University noted that mitochondria are known to release dsRNA into the cytoplasm in response to stress. “Because of this, it is not uncommon to observe an IFN response in the face of cell toxicity,” he wrote. “Regardless of the source of the cdsRNA, its presence can be problematic.” Wolozin considers the finding that JAK inhibitors rescued toxicity from cdsRNA an important contribution to the field, as it raises the question of whether brain-penetrant JAK inhibitors might treat C9ORF72 ALS/FTD.
Albers told Alzforum that plans are underway for a clinical trial that will evaluate baricitinib in people with ALS or AD. He expects the basket trial—one testing the same drug in different diseases—will begin in late 2021.
Freibaum, of St. Jude’s, noted that the study dovetails with recent findings implicating the RNA helicase DDX3X in C9ORF72 toxicity. The helicase unravels secondary structures formed by repeat-laden C9ORF72 transcripts, readying them for RAN translation and promoting the formation of dipeptide repeats. Freibaum noted that such helicases become trapped in liquid condensates along with C9ORF72 dipeptides, TDP-43, and other RNA-binding proteins, and wondered whether the snagging of such helicases might promote dsRNA formation (Cheng et al., 2019; Tseng et al., 2021).
Sami Barmada of the University of Michigan, Ann Arbor, brought up another pathway by which C9ORF72 expansions might promote accumulation of RNA species including dsRNA. The expansions douse expression of C9ORF72, which functions in autophagy. This loss is thought to speed up accumulation of aggregation-prone protein products of the expansions, i.e., dipeptide repeats, but it could also trigger a build-up of unwanted RNA species themselves, as these are also degraded via autophagy (Frankel et al., 2017).
The propagation of cdsRNA, and the ensuing IFN response, evokes the mechanisms by which cells handle dsRNA derived from viruses such as HSV-1. Via the receptor Sidt2, cells take up extracellular dsRNA into the cytoplasm, a move that is required to instigate the IFN response (Nguyen et al., 2017). The idea that reactivation of latent viruses, including HSV-1, can kick off or accelerate AD pathogenesis has gathered steam in recent years, and now genomic dsRNAs may be among the potential culprits, as well (Jun 2018 news).—Jessica Shugart
References
News Citations
- It’s ‘And,’ Not ‘Either-Or’: C9ORF72 Mechanisms of Action are Linked
- Herpes Triggers Amyloid—Could This Virus Fuel Alzheimer’s?
Paper Citations
- Guimarães PO, Quirk D, Furtado RH, Maia LN, Saraiva JF, Antunes MO, Kalil Filho R, Junior VM, Soeiro AM, Tognon AP, Veiga VC, Martins PA, Moia DD, Sampaio BS, Assis SR, Soares RV, Piano LP, Castilho K, Momesso RG, Monfardini F, Guimarães HP, Ponce de Leon D, Dulcine M, Pinheiro MR, Gunay LM, Deuring JJ, Rizzo LV, Koncz T, Berwanger O, STOP-COVID Trial Investigators. Tofacitinib in Patients Hospitalized with Covid-19 Pneumonia. N Engl J Med. 2021 Jun 16; PubMed.
- Saldi TK, Ash PE, Wilson G, Gonzales P, Garrido-Lecca A, Roberts CM, Dostal V, Gendron TF, Stein LD, Blumenthal T, Petrucelli L, Link CD. TDP-1, the Caenorhabditis elegans ortholog of TDP-43, limits the accumulation of double-stranded RNA. EMBO J. 2014 Dec 17;33(24):2947-66. Epub 2014 Nov 12 PubMed.
- Tam OH, Rozhkov NV, Shaw R, Kim D, Hubbard I, Fennessey S, Propp N, NYGC ALS Consortium, Fagegaltier D, Harris BT, Ostrow LW, Phatnani H, Ravits J, Dubnau J, Gale Hammell M. Postmortem Cortex Samples Identify Distinct Molecular Subtypes of ALS: Retrotransposon Activation, Oxidative Stress, and Activated Glia. Cell Rep. 2019 Oct 29;29(5):1164-1177.e5. PubMed.
- Cheng W, Wang S, Zhang Z, Morgens DW, Hayes LR, Lee S, Portz B, Xie Y, Nguyen BV, Haney MS, Yan S, Dong D, Coyne AN, Yang J, Xian F, Cleveland DW, Qiu Z, Rothstein JD, Shorter J, Gao FB, Bassik MC, Sun S. CRISPR-Cas9 Screens Identify the RNA Helicase DDX3X as a Repressor of C9ORF72 (GGGGCC)n Repeat-Associated Non-AUG Translation. Neuron. 2019 Dec 4;104(5):885-898.e8. Epub 2019 Oct 3 PubMed.
- Tseng YJ, Sandwith SN, Green KM, Chambers AE, Krans A, Raimer HM, Sharlow ME, Reisinger MA, Richardson AE, Routh ED, Smaldino MA, Wang YH, Vaughn JP, Todd PK, Smaldino PJ. The RNA helicase DHX36-G4R1 modulates C9orf72 GGGGCC hexanucleotide repeat-associated translation. J Biol Chem. 2021 Aug;297(2):100914. Epub 2021 Jun 24 PubMed.
- Frankel LB, Lubas M, Lund AH. Emerging connections between RNA and autophagy. Autophagy. 2017 Jan 2;13(1):3-23. Epub 2016 Oct 7 PubMed.
- Nguyen TA, Smith BR, Tate MD, Belz GT, Barrios MH, Elgass KD, Weisman AS, Baker PJ, Preston SP, Whitehead L, Garnham A, Lundie RJ, Smyth GK, Pellegrini M, O'Keeffe M, Wicks IP, Masters SL, Hunter CP, Pang KC. SIDT2 Transports Extracellular dsRNA into the Cytoplasm for Innate Immune Recognition. Immunity. 2017 Sep 19;47(3):498-509.e6. Epub 2017 Sep 12 PubMed.
Further Reading
No Available Further Reading
Primary Papers
- Rodriguez S, Sahin A, Schrank BR, Al-Lawati H, Costantino I, Benz E, Fard D, Albers AD, Cao L, Gomez AC, Evans K, Ratti E, Cudkowicz M, Frosch MP, Talkowski M, Sorger PK, Hyman BT, Albers MW. Genome-encoded cytoplasmic double-stranded RNAs, found in C9ORF72 ALS-FTD brain, propagate neuronal loss. Sci Transl Med. 2021 Jul 7;13(601) PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Boston University School of Medicine
The work from the Albers group presents an additional mechanism through which the hexanucleotide expansions of C9ORF72 can mediate cell death. The group began by studying motor cortices (and frontal cortices) from ALS/FTD C9ORF72 patients and observed the presence of cytoplasmic double-stranded RNA (cdsRNA) that co-localized with TDP-43 pathology. This data suggests that TDP-43 pathology correlates with cdsRNA, but does not show the converse because it is unclear what fraction of cells had cdsRNA pathology but no TDP-43 pathology.
Immunoblotting and transcriptome analysis suggested that the cdsRNA induced an IFN-I response, which is consistent with known responses to cdsRNA. The group went on to show that expression of 66mer G4C2 repeat sequences induces a similar IFN response and some cell death; the response depended on the presence of cdsRNA but could be elicited with cdsRNA from transcripts unrelated to G4C2, such as GFP cdsRNA.
The lack of specificity for G4C2 is an important caveat, because mitochondrial damage is known to release dsRNA into the cytoplasm, creating cdsRNA. Because of this, it is not uncommon to observe an IFN response in the face of cell toxicity.
Regardless of the source of the cdsRNA, its presence can be problematic. The Albers group addressed this by inhibiting different kinases regulating the IFN pathway. They examined PKR inhibitors, but found those to be toxic. On the other hand, JAK inhibitors nicely rescued the toxicity. This raises the possibility that brain-penetrant JAK inhibitors might have some value in treating C9ORF72 ALS/FTD, which is a potentially important contribution.
Make a Comment
To make a comment you must login or register.