Drug and Biomarker Candidates for C9ORF72 ALS and FTD
Quick Links
In the August 14 Neuron, researchers offer up a possible treatment and a potential biomarker for amyotrophic lateral sclerosis and frontotemporal dementia caused by mutations in the C9ORF72 gene. Leonard Petrucelli at the Mayo Clinic in Jacksonville, Florida, and Matthew Disney at The Scripps Research Institute in Jupiter, Florida, developed small molecules that bind to the RNA produced by the malignant gene. The compounds stick to GGGGCC RNA repeats and prevent their translation into polypeptides. The authors also reported that one of those peptides, poly(glycine-proline), accumulates in the cerebrospinal fluid (CSF) of C9ORF72 expansion carriers, making it a candidate biomarker to quickly evaluate whether C9ORF72-targeted therapies work in people.
Between the compounds and the biomarker, the paper has excellent potential to contribute to future clinical studies, commented Nicholas Maragakis of the Johns Hopkins University School of Medicine in Baltimore, who was not involved in the study.
Binding RNA Repeats
Long hexanucleotide repeats in intron 1 of the C9ORF72 gene are the most common genetic cause of ALS and FTD. Most people carry a few of the GGGGCC repeats, but some have hundreds or thousands, and this predisposes them to disease (see Sep 2011 news story). The corrupt gene produces repetitive RNAs going in both the sense (GGGGCC) and antisense (CCCCGG) direction, and these form foci that researchers suspect may be toxic (see Oct 2013 news story). Moreover, those RNAs undergo nonstandard translation, creating five different dipeptide products, which might also cause disease (see Feb 2013 news story; news story).
In this study, Disney and Petrucelli took aim at both the repeat RNAs and peptides with small molecules that bind the RNAs and block translation. Disney had previously developed a small molecule that binds to CGG repeats in the fragile X mental retardation 1 (FMR1) gene, which cause fragile X-associated tremor ataxia syndrome. The scientists called the drug 1a for simplicity; the full name of this five-ring molecule is 9-hydroxy-5,11-dimethyl-2-(2-(piperidin-1-yl)ethyl)-6H-pyrido[4,3-b]carbozol-2-ium. Disney wondered if 1a might also interact with the GGGGCC sequences in the C9ORF72 repeat expansion, and approached Petrucelli to collaborate.
Zhaoming Su in Disney’s lab, one of the study’s first authors, screened 132 molecules that had structures reminiscent of 1a’s and might bind RNA. This led him to three lead compounds that interacted with the C9ORF72 repeats, 1a and the repeats the authors named 2 and 3. In Petrucelli’s group, co-first authors Yongjie Zhang, Tania Gendron, and Peter Bauer tested the three compounds in human embryonic kidney (HEK) cultures that expressed 66 GGGGCC repeats. This RNA forms foci in the cells, but treatment with either 1a or 2 decreased their number (see image below). The HEK cells translated the RNAs into dipeptides poly(GP) and poly(glycine-alanine), but 1a and 2 knocked down production of those peptides. Compound 3, in contrast, did not affect RNA foci and only mildly inhibited poly(GP) production.
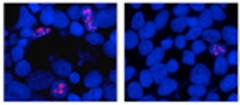
C9ORF72 repeats cause RNA foci (pink, left) to form in cultured cells, but treatment with compound 1a (right) reduces foci numbers. [Image courtesy of Neuron, Su et al.]
Next, Petrucelli’s group tested compounds 1a and 2 in a cell model that more closely mimics the neurons affected in FTD and ALS. They obtained fibroblasts from people with the C9ORF72 repeat expansion and differentiated them into induced neurons (iNeurons; see Jan 2013 news story). Compound 2 was toxic to these cells, but 1a reduced both RNA foci and poly(GP).
Disney and colleagues continue to screen for even better compounds. Petrucelli hopes to test 1a in other patient cell lines and in animal models, which are under development for C9ORF72 expansions. While 1a crosses the blood-brain barrier, it interacts only with repeats going in the sense direction. Disney wants to develop a small molecule that could target the antisense RNA, too, or possibly both repeat RNAs at once.
Peptide Biomarker
Should Disney’s or any other C9ORF72-specific treatment (see Oct 2013 news story) reach the clinical trial stage, researchers would benefit from a quick test of whether the drug works. Petrucelli’s group has developed an antibody-based assay to identify one of the repeat peptides, poly(GP), in cerebrospinal fluid. They tested the assay on samples from 14 people with ALS due to C9ORF72 expansions, 25 people with ALS but no expansions, and five healthy controls. The people with expansions clearly had more poly(GP) than the other two groups. “It was pretty remarkable separation between the healthy people and the C9ORF72 patients,” commented Robert Baloh of Cedars-Sinai Medical Center in Los Angeles, who did not participate in the work. “That is almost a diagnostic test, let alone a biomarker.” He said the biomarker could be a “game-changer” for clinical trials, allowing researchers to speedily assess efficacy.
Among people with the C9ORF72 expansion, poly(GP) levels varied more than 10-fold. This raises many further questions about how CSF dipeptide concentrations might correlate with repeat number, disease onset, disease severity, or other factors. Petrucelli plans to examine the biomarker further in a longitudinal study. His group also aims to develop assays against the four other dipeptide repeats made by C9ORF72 expanded RNAs.—Amber Dance
References
News Citations
- Corrupt Code: DNA Repeats Are Common Cause for ALS and FTD
- RNA Deposits Confer Toxicity in C9ORF72 ALS
- RNA Twist: C9ORF72 Intron Expansion Makes Aggregating Protein
- Second Study Sees Intron in FTLD Gene Translated
- Stem Cells: Simpler to Make, Easier on the Immune System
- Second Study Confirms Antisense Oligonucleotides Bust RNA Aggregates
Further Reading
Papers
- Zamiri B, Reddy K, Macgregor RB Jr, Pearson CE. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J Biol Chem. 2014 Feb 21;289(8):4653-9. Epub 2013 Dec 26 PubMed.
- Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014 Mar;127(3):359-76. Epub 2014 Jan 7 PubMed.
- Simón-Sánchez J, Dopper EG, Cohn-Hokke PE, Hukema RK, Nicolaou N, Seelaar H, de Graaf JR, de Koning I, van Schoor NM, Deeg DJ, Smits M, Raaphorst J, van den Berg LH, Schelhaas HJ, De Die-Smulders CE, Majoor-Krakauer D, Rozemuller AJ, Willemsen R, Pijnenburg YA, Heutink P, van Swieten JC. The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain. 2012 Mar;135(Pt 3):723-35. PubMed.
- Gendron TF, Cosio DM, Petrucelli L. c9RAN translation: a potential therapeutic target for the treatment of amyotrophic lateral sclerosis and frontotemporal dementia. Expert Opin Ther Targets. 2013 Sep;17(9):991-5. PubMed.
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013 Feb 20;77(4):639-46. PubMed.
News
- C9ORF72’s Dirty Work Done by Problem Proteins
- C9ORF72 Killer Dipeptides Clog the Nucleolus
- C9ORF72 Repeats Expand into New Disorders—Cause, or Coincidence?
- RNA-DNA Pairs: At the Root of C9ORF72 Repeat Damage?
- Researchers Revel in C9ORF72 Advances at RNA Symposium
- Methylation a Turn Off for Disease Gene C9ORF72?
Primary Papers
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, Johnston A, Overstreet K, Oh SY, Todd PK, Berry JD, Cudkowicz ME, Boeve BF, Dickson D, Floeter MK, Traynor BJ, Morelli C, Ratti A, Silani V, Rademakers R, Brown RH, Rothstein JD, Boylan KB, Petrucelli L, Disney MD. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014 Sep 3;83(5):1043-50. Epub 2014 Aug 14 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.