Scientists Hunting Rare Alzheimer’s Mutations Eye Phospholipase D3
Quick Links
Having surveyed the genome for common variants associated with Alzheimer’s disease, geneticists are now turning the search lights on rare, hard-to-find mutations that may be even stronger risk factors. At the Alzheimer’s Association International Conference (AAIC), held July 13-18 in Boston, researchers presented potential, if unconfirmed, candidates, including phospholipase D3, the ubiquitin ligase TTC3, and genes that play roles in axon guidance and microtubule stabilization. Scientists report some success sequencing the exomes, or protein-coding regions, of genes from families hard hit with AD. They are thus far having less luck with a separate method called exome chip technology. It searches for rare variants by skimming the genome’s surface, genotyping specific, known mutations in large numbers of people. Researchers at AAIC agreed that at this point, identifying and validating rare variants remains challenging.
Nonetheless, this approach is slowly beginning to pay dividends. Last year, two research groups discovered that rare mutations in the immune cell receptor TREM2 confer as much risk for AD as ApoE4 (see Alzforum News story on Guerreiro et al., 2013 and Jonsson et al., 2013). None of the new variants discussed at AAIC have yet reached TREM2’s status as a definite risk factor, commented Adam Naj of the University of Pennsylvania in Philadelphia. “Everybody was presenting preliminary work that needs a lot of replication,” said Naj, who co-chaired one of two sessions on rare variants at the conference.
Researchers are taking two main approaches to find these rare variants. Some identify families where AD affects several members, and then completely sequence their exomes to find variants that co-segregate with the disease. Others genotype large groups of cases and controls with exome chips that identify known single nucleotide polymorphisms (SNPs). The chip approach saves money compared to deep sequencing many samples, but it cannot identify variants beyond the known SNPs imprinted on the chip. To date, the family approach looks more promising, said Carlos Cruchaga of Washington University in St. Louis, Missouri. One reason is that exome chips still have some technical problems, including high rates of false positive and false negative hits.
Whichever method they choose, researchers looking for rare variants have to do a lot of sifting to find the truly interesting ones. Each person totes thousands of SNPs, and the first-pass analysis will likely identify several that appear linked to disease. “The problem is not finding rare SNPs, it is figuring out which ones are actually doing anything,” said Mark Logue of Boston University. “The trick is prioritizing them for follow-up.” Scientists can do this by checking whether a mutation is likely to have functional consequences, for example if it alters protein structure, or if the nucleotide in question is conserved in evolution, which would indicate an important role in biology.
Debuting Candidates
Here is a taste of the preliminary findings discussed in Boston:
Performing exome sequencing on 14 families, Cruchaga’s team turned up a variant in the phospholipase D3 gene. It appeared in people with Alzheimer’s from two of the clans. In further genotyping of 10,000 cases and controls, the researchers confirmed the variant associated with AD. Sequencing of the gene in a further 4,000 individuals turned up other mutations in the gene that may affect AD risk. Cruchaga suspects that phospholipase D3 could turn out to have as strong an effect as TREM2. However, little is known about the protein’s function, he told Alzforum.
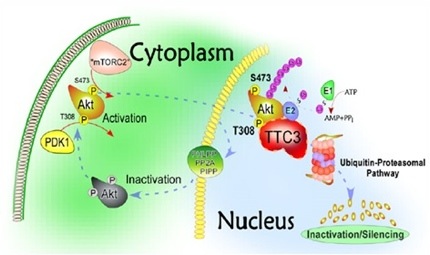
Martin Kohli, who works with Peggy Pericak-Vance at the University of Miami, Florida, presented another family-based study. One of six kindreds examined led him to mutations in the gene tetratricopeptide repeat domain 3. TTC3 is a ubiquitin protein ligase that is crucial for neuron survival (see image below). The AD-linked variant was absent from 10,000 control alleles. Pericak-Vance found the candidate intriguing because of its function in survival signaling pathways, which could easily be linked to dementia. Moreover, it is located in the “Down’s syndrome critical region” on chromosome 21, Pericak-Vance said. Some people with Down’s syndrome have a triple dose of not the entire chromosome 21, but just this fragment. Because most people with Down’s syndrome develop Alzheimer’s, researchers had suspected that some AD-linked gene was hiding in that critical region, Pericak-Vance said. She suggested TTC3 might be it. APP resides outside of this region on chromosome 21, but the critical region includes the BACE2 gene.
TTC3 helps degrade proteins.
[Image courtesy Martin Kohli, University of Miami, Florida, and Developmental Cell]
Robert Graham of Genentech, in South San Francisco, California, reported on exome and whole-genome sequencing of another extended family. This project identified a rare variant that Graham said was more likely to occur in cases than controls in a separate, larger series of subjects. This mutation lies in the gene unc-5 homolog C. UNC5c is one of a family of receptors for netrins. Netrins work during neural development to regulate axon guidance. The researchers found that the mutation reduced cell viability in vitro.
Other researchers have adopted different strategies. Logue, who works in Lindsay Farrer’s laboratory at Boston University, chose to study an ethnic group, that is, African Americans. This population has been under-studied and could offer new insight into genetic risk factors, Logue said. He began by sequencing the exomes of eight African Americans who have a family history of AD. From there, he identified 63 prime suspect SNPs, which he genotyped in a further 435 cases and 422 controls. He discovered 11 cases who had one or two variants in a gene called A kinase anchor protein 9 (AKAP9). One control possessed a rare variant in AKAP9 as well. The AKAP9 protein participates in microtubule stabilization, as does the AD-related protein tau.
The Alzheimer’s Disease Genetics Consortium (ADGC) has pursued the exome chip approach. Li-San Wang of the University of Pennsylvania, Philadelphia, reported that after more than 16,000 subjects having been genotyped, no novel variant has yet reached statistical significance for an association with AD. A larger sample will have to be tested, suggested Wang. One interesting candidate was mucin 17 (MUC17), Naj told Alzforum. Mucins decorate the membranes of epithelial cells where they participate in structure maintenance and signaling.
Rare Findings
Going forward, geneticists will continue to be hampered by the very rarity of what they seek. When scientists find a SNP in a gene in a single family, how can they confirm its association with disease? It might require tens of thousands of individuals to find another with that same variant. The SNP might even be a “private mutation,” unique to one individual or kindred.
Scientists predict they can get around this difficulty by looking for cases of multiple variants in the same gene. In addition, if they cannot find more people with the variant of interest, they can turn to the lab and study the gene in cells and models systems, looking for clues that it might contribute to brain function.
Because it takes so many subjects to find and confirm rare SNPs, scientists are working together in large collaborations. For example, the Alzheimer’s Disease Neuroimaging Initiative (ADNI) is performing whole-genome sequencing, which should be completed imminently, said Robert Green, Harvard University, who works on the project. Members of the International Genomics of Alzheimer’s Project (IGAP) plan to pool their exome chip data. Part of the 2012 Presidential Initiative against AD (see ARF related news story) includes funds for the Alzheimer’s Disease Sequencing Project (ADSP). The project’s first phase, already underway, is to sequence the genome of 582 people from 111 families with high rates of AD. Researchers expect the earliest results from this venture in the next year. In its second phase, ADSP will sequence the exomes of 5,000 AD cases and 5,000 controls.
Cruchaga, Logue, and Wangall predicted plenty more rare variant news in the next few years. Alison Goate of Washington University in St. Louis, Missouri, summed up the attitude of AD geneticists. “Time will tell as to whether any of these findings prove to be replicable and real associations,” she told Alzforum.—Amber Dance.
References
News Citations
- Enter the New Alzheimer’s Gene: TREM2 Variant Triples Risk
- New Shot in Arm for Alzheimer's Research Announced Today
Paper Citations
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J, Alzheimer Genetic Analysis Group. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013 Jan 10;368(2):117-27. Epub 2012 Nov 14 PubMed.
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013 Jan 10;368(2):107-16. Epub 2012 Nov 14 PubMed.
Other Citations
External Citations
Further Reading
Papers
- Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, Jiménez-Velazquez IZ, Rogaeva E, St George-Hyslop PH, Mayeux R. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol. 2011 Mar;68(3):320-8. PubMed.
- Zimprich A, Benet-Pagès A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, Rossle SC, Klopp N, Wolf E, Seppi K, Pirker W, Presslauer S, Mollenhauer B, Katzenschlager R, Foki T, Hotzy C, Reinthaler E, Harutyunyan A, Kralovics R, Peters A, Zimprich F, Brücke T, Poewe W, Auff E, Trenkwalder C, Rost B, Ransmayr G, Winkelmann J, Meitinger T, Strom TM. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011 Jul 15;89(1):168-75. PubMed.
- Shulman JM, Chipendo P, Chibnik LB, Aubin C, Tran D, Keenan BT, Kramer PL, Schneider JA, Bennett DA, Feany MB, De Jager PL. Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am J Hum Genet. 2011 Feb 11;88(2):232-8. PubMed.
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O'Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012 Sep 6;489(7414):91-100. PubMed.
News
- Thousands of Whole Genomes to Be Mined for New Clues to AD
- Coverage Denial For Amyloid Scans Riles Alzheimer’s Community
- Pooled GWAS Reveals New Alzheimer’s Genes and Pathways
- Dementia Prevalence Falls in England
- Are Subtle Memory Concerns a Sign of Future Dementia?
- Clinical Trials Roundup: Four Hits, No Home Runs
- Clinical Trials Roundup: Broadening the Lines of Attack
- Are Protein Strains The Cause of Different Tauopathies?
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.