Modern Microscopy Plumbs the Depths of Brain Tissue
Quick Links
Despite its popularity, live imaging of the brain has a drawback: It is difficult to go beyond the upper layers of the cortex (see Part 1). To reach its depths, scientists generally must excise brain slices or fix tissue. This type of imaging has its own challenges, but two recent papers rise to meet them. For one, researchers at TissueVision, Inc., of Cambridge, Massachusetts, and the Cold Spring Harbor Laboratory on Long Island, New York, have developed a machine that automatically sections and scans an entire small animal brain, all while scientists are busy elsewhere (or out for coffee). In the January 15 Nature Methods, the team explained the technology, which is already hard at work on a large-scale project by the Allen Institute for Brain Science in Seattle, Washington. Another difficulty is that the ability of microscopes to scan and refocus in the z-direction (i.e., perpendicular to the plane of a tissue slice) is currently too slow to watch fast events such as neural signaling. On that one, researchers at the University of Oxford, U.K., found a trick to speed up z-scanning without sacrificing image quality. The scientists nudge a mirror instead of the objective lens. They reported their finding in the February 12 Proceedings of the National Academy of Sciences.
Mirror, Mirror….
The slow rate of z-scanning has been a major limitation of multiphoton microscopy, wrote first author Edward Botcherby, senior author Tony Wilson, and colleagues. The team custom-built a microscope to quicken things. They first described the concept in 2007 (Botcherby et al., 2007). In the current work they apply it to neuroscience, where speed is necessary to catch, for example, the flashes of calcium that indicate neural activity. The average calcium wave moves at 15-30 microns per second at room temperature (Jaffe, 1993). With their new system, Botcherby and colleagues could go up or down by more than 60 microns in a millisecond.
The new microscope comprises not only a lateral scan unit for the xy-direction, but an additional, novel axial scan unit for the z-direction. The axial unit includes a second lens and a mirror; it is the mirror’s movement that refocuses the laser beam. The lens and mirror work together such that all light rays enter the desired focal point, keeping the image crisp. Since the actual sample remains motionless, movement artifacts are limited.
To test the system, Botcherby successfully monitored the calcium-sensitive dyes Fluo-4 and Alexa 594 lighting up dendritic activity in mouse brain slabs and slices after signal induction. “The system we have developed will provide neuroscientists with the tools necessary to study neuronal activity at the population and subcellular level over large enough distances and at a speed that allows the activity of single-action potentials to be followed,” the authors concluded. “This will enable us to study synaptic integration and the neuronal population activity at an unprecedented level of detail.”
Other researchers have developed different technologies for z-scanning (Duemani Reddy et al., 2008; Grewe et al., 2010; Saggau, 2006; Kirkby et al., 2010). However, “this is the way to go. Simple and fast,” said David Kleinfeld of the University of California, San Diego, who was not involved in the project.
Look Ma, No Hands
To obtain a 3D picture of the brain has long required that scientists reduce the sample to a series of 2D slices. The process of mounting and cleaving the brain of a mouse, individually imaging each slice, and reassembling the images into a single digital dataset can take weeks if done by hand, said Pavel Osten of Cold Spring Harbor Laboratory, an author on the Nature Methods paper. In addition, the physical slicing can mar or tear the valuable sample, making it difficult to precisely realign the images of each slice. Osten’s coauthor, Tim Ragan at TissueVision, came up with a better way, and it takes as few as four hours, Ragan said. “We are trying to transform the field of histology from something that has been a very tedious, labor-intensive endeavor to something that is going to be high throughput,” Ragan said. “We are industrializing histology.”
Ragan and Osten call their system serial two-photon tomography, or STP tomography. The only hands-on part is when users fix a whole, fluorescently labeled brain and mount it in an agar block. They place this under the microscope, push the start button, and walk away. The microscope then performs two steps serially. First, it uses two-photon technology to image the area just beneath the top of the sample. Second, it uses a vibratome to slice off the top. Any damage done by the blade will not be part of the final dataset. “We always image the tissue before we take the section,” Ragan said. After some 260 image-slice repetitions, the system reassembles the complete 3D image. Ragan tested the microscope with mouse brains expressing transgenes for green fluorescent protein (GFP) in neurons or oligodendrocytes.
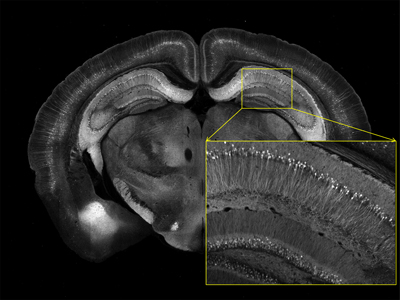
Serial Two-Photon Tomography of Mouse Brain
This image was taken with STP tomography. Neurons express yellow fluorescent protein. Image credit: Tim Ragan, TissueVision, Inc., Cambridge, Massachusetts
The technology can be used with fluorescent tracers to label neural projections from one part of the brain to another. In two test cases, the authors injected the retrograde tracer cholera toxin B subunit-Alexa Fluor 488 or the anterograde tracer adeno-associated virus expressing GFP into mouse brains to track neural connections. They observed previously unknown connections, including a link between the somatosensory barrel cortex and the contralateral orbital cortex, and a junction between the same barrel cortex and the contralateral motor cortex. These anatomical links are sparse, and the researchers do not know what they mean for neurophysiology, yet they showcase how sensitive the technique is, Osten said.
Moreover, STP tomography may reveal when the brain responds to stimuli. Osten plans to image fluorescent markers for immediate early genes indicating neuronal activation, such as c-fos and Arc. By fixing the brain after an animal receives a drug, say, or performs a behavior, scientists could start to figure out which neurons are activated at that time. The immediate early genes are expressed about three hours after a stimulus, Osten said, so there is ample time to extract and fix the brain. This method, he said, could be used to study learning and memory, or to understand the brains of Alzheimer’s and Parkinson’s model animals. Osten plans to image and compare the brains of diverse mouse models from schizophrenia to autism.
STP tomography is a perfect tool for researchers on the Allen Mouse Brain Connectivity Atlas, said Hongkui Zeng, senior scientific director for research and development at the Allen Institute. Over the next couple of years, she and colleagues plan to put together the wiring diagram for a mouse brain with the help of six TissueVision microscopes. “We are very happy with the system,” Zeng said. “No sectioning is needed anymore, saving a lot of labor.”
“The biggest advantage of the system is that it eliminates the need for mounting and dehydration of the tissue sections,” wrote Ahmad Salehi of the Palo Alto Veterans Affairs Health Care System in California, in an e-mail to ARF (see full comment below). Salehi, who was not involved in the TissueVision study, is furthermore taking advantage of modern computing power to stitch together images digitally instead of manually. He takes dozens of images of different pieces of the brain and lets the computer join them to form one big, seamless picture. He has used the technique on the dentate gyrus, but said it could work for any region (see ARF related news story).
With that technique, scientists still have to take the images themselves. Using even newer technology, Salehi leaves that bit to machines as well. His NanoZoomer from Hamamatsu Photonics, K.K., of Hamamatsu City in Japan, can automatically image a slide—a tissue slice, for example—in half an hour. It then renders a single image that scientists can dig into, rather like zooming in on your house in Google Maps. “I think in a few years there will no longer be any need for conventional microscopy, steering, and collecting images piece by piece,” Salehi told ARF.
How Low Can You Go?
For now, deep-tissue imaging is mostly limited to ex-vivo samples, while live-animal imaging is mostly limited to shallow depths (see Part 1). However, scientists are pushing the depth limit for living tissue. Chris Xu at Cornell University in Ithaca, New York, managed to image 1.6 millimeters deep into the cortex of a living mouse—“currently the world record,” he said—using two-photon microscopy with 1,300-nanometer infrared light. However, this works only with red fluorophores, and the image becomes faint as he approaches the depth limit (Kobat et al., 2011).
Other researchers apply invasive means to see deeper. For example, Michael Levene at Yale University uses a prism to bounce light waves below the surface, giving him a field of view approximately 900 microns deep (Chia and Levene, 2009). “It is conceptually straightforward,” Levene said, admitting that pushing a prism into the brain’s surface is rather crude. “It is like a little periscope, so it lets us image side-on to the tissue instead of top-down,” he said. In this way, Levene can observe all six layers of the cortex in one picture. In contrast, most in-vivo imaging techniques only offer a view of planes parallel to the organ’s surface, one cortical layer at a time.
Endoscopes are another useful, albeit invasive, tool to dig deeper than light waves allow (Rivera et al., 2011; see ARF related news story on Barretto et al., 2011).
“Deep tissue imaging is a very exciting field,” Xu said. “Now you can go millimeters to centimeters…. We can now see the brain areas that no one else was able to look at before, with high resolution.”—Amber Dance
References
News Citations
- Modern Microscopy Skims Surface of Living Minds and Spines
- Computer-Painted Pictures Worth a Thousand Words
- Reel World—Viewing Long-Term Changes in Neuron Structure, Activity
Paper Citations
- Botcherby EJ, Juskaitis R, Booth MJ, Wilson T. Aberration-free optical refocusing in high numerical aperture microscopy. Opt Lett. 2007 Jul 15;32(14):2007-9. PubMed.
- Jaffe LF. Classes and mechanisms of calcium waves. Cell Calcium. 1993 Nov;14(10):736-45. PubMed.
- Duemani Reddy G, Kelleher K, Fink R, Saggau P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat Neurosci. 2008 Jun;11(6):713-20. PubMed.
- Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat Methods. 2010 May;7(5):399-405. PubMed.
- Saggau P. New methods and uses for fast optical scanning. Curr Opin Neurobiol. 2006 Oct;16(5):543-50. PubMed.
- Kirkby PA, Srinivas Nadella KM, Silver RA. A compact Acousto-Optic Lens for 2D and 3D femtosecond based 2-photon microscopy. Opt Express. 2010 Jun 21;18(13):13721-45. PubMed.
- Kobat D, Horton NG, Xu C. In vivo two-photon microscopy to 1.6-mm depth in mouse cortex. J Biomed Opt. 2011 Oct;16(10):106014. PubMed.
- Chia TH, Levene MJ. Microprisms for in vivo multilayer cortical imaging. J Neurophysiol. 2009 Aug;102(2):1310-4. PubMed.
- Rivera DR, Brown CM, Ouzounov DG, Pavlova I, Kobat D, Webb WW, Xu C. Compact and flexible raster scanning multiphoton endoscope capable of imaging unstained tissue. Proc Natl Acad Sci U S A. 2011 Oct 25;108(43):17598-603. PubMed.
- Barretto RP, Ko TH, Jung JC, Wang TJ, Capps G, Waters AC, Ziv Y, Attardo A, Recht L, Schnitzer MJ. Time-lapse imaging of disease progression in deep brain areas using fluorescence microendoscopy. Nat Med. 2011 Feb;17(2):223-8. PubMed.
Further Reading
Papers
- Xie T, He Y. Mapping the Alzheimer's brain with connectomics. Front Psychiatry. 2011;2:77. PubMed.
- Martin G. Network analysis and the connectopathies: current research and future approaches. Nonlinear Dynamics Psychol Life Sci. 2012 Jan;16(1):79-90. PubMed.
- Sporns O. The human connectome: a complex network. Ann N Y Acad Sci. 2011 Apr;1224:109-25. PubMed.
- Landahl EC, Antipova O, Bongaarts A, Barrea R, Berry R, Binder LI, Irving T, Orgel J, Vana L, Rice SE. X-ray diffraction from intact tau aggregates in human brain tissue. Nucl Instrum Methods Phys Res A. 2011 Sep 1;649(1):184-187. PubMed.
- Garbuzova-Davis S, Haller E, Saporta S, Kolomey I, Nicosia SV, Sanberg PR. Ultrastructure of blood-brain barrier and blood-spinal cord barrier in SOD1 mice modeling ALS. Brain Res. 2007 Jul 9;1157:126-37. PubMed.
News
- Miami: Multimodal Imaging, New Way to Test Amyloid Hypothesis
- Enabling Technologies for Alzheimer Disease Research: Seventh Bar Harbor Workshop, 2007, Part 4
- Computer-Painted Pictures Worth a Thousand Words
- Two-Photon Microscopy Spies on Single Synapses
- Reel World—Viewing Long-Term Changes in Neuron Structure, Activity
- Modern Microscopy Skims Surface of Living Minds and Spines
Primary Papers
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012 Mar;9(3):255-8. PubMed.
- Botcherby EJ, Smith CW, Kohl MM, Débarre D, Booth MJ, Juškaitis R, Paulsen O, Wilson T. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):2919-24. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Stanford University School of Medicine, VA Palo Alto Health Care System
In any 3D reconstruction method compiled from images captured from mounted sections, correction for differential positioning of each tissue section and non-homogeneous dehydration of various brain regions are the prime concerns. I think the biggest advantage of the TissueVision system is that it completely eliminates the need for mounting and dehydration of the tissue sections after cutting. Although I am a bit concerned that mechanical sectioning of brains with vibratome may deform and reposition the tissue samples, it appears that the authors are confident that covalent crosslinking with agarose, in which the brain is embedded, would solve this potential problem. In summary, the system is another big step toward partially replacing conventional histological methods in high-throughput analysis of the entire brain of rodent models of different diseases.
Make a Comment
To make a comment you must login or register.