Thanks for the Memories—KIBRA Alleles Predict Top Memory Performers
Quick Links
20 October 2006. If you are one of those people who can recall verbatim a conversation held a year ago, then you might want to thank your parents—not necessarily for sending you to that prestigious school, forcing you to memorize all of the Canterbury Tales, or spending endless hours playing The Memory Game, but for simply passing on a specific letter of the genetic code. A paper in today’s Science reports that a single allelic variation (T for C) in the KIBRA gene can increase memory performance by about 20 percent. Furthermore, non-carriers of the T allele show greater neuronal activation in the hippocampus and parahippocampal gyrus, brain centers that are intimately involved in learning and memory, suggesting that they have to work harder when remembering. The findings could lead to new strategies to develop memory-enhancing drugs that might help patients with mild cognitive impairment or even those with Alzheimer disease and other dementias.
The KIBRA gene codes for a protein that has binding motifs for dendrin and a protein kinase C isoform called PKMζ, both linked to long-term potentiation and synaptic plasticity (see Pastalkova et al. 2006, ARF related news story, and Kremerskothen et al., 2003)—processes that are essential for human learning and memory. But it was not these protein-protein interactions that led to the discovery of the memory allele. Instead, Andreas Papassotiropoulos and Dominique de Quervain at the University of Zurich, Switzerland, Dietrich Stephan at the Translational Genomics Research Institute, Phoenix, Arizona, and their colleagues found the KIBRA allele in an unbiased screen for genetic variations that impact memory.
The authors compared a genome-wide screen of more than half a million single nucleotide polymorphisms (SNPs) with performance in a verbal memory task. They recruited 351 young adults (mean age 22 years) from Switzerland to take part in the screen, testing their verbal episodic memory by having them remember 30 unrelated nouns. The authors found that two SNPs were associated with volunteers with the best memories: rs17070145, a T-to-C substitution in intron 9 of the KIBRA gene; and rs643988, another T-to-C substitution in the first intron of the gene encoding the synaptic protein calsyntenin 2 (CLSTN2). Volunteers with the T variant of the KIBRA gene had 24 percent better recall 5 minutes after being presented with the words and 19 percent better recall after 24 hours. These differences were highly significant. The differences between those with the two CLSTN alleles were not as high—16 and 15 percent for the short and long recall, respectively.
Genetic association studies in large diverse populations can be notoriously difficult to interpret because the cohort being studied may not necessarily be genetically similar. So the authors verified the findings in two ways. First, to eliminate the possibility of false positives, they checked for heterogeneity within the study sample, finding that the volunteers were genetically homogeneous apart from 10 outliers who were discounted from the study. Second, they performed the same analysis on an American cohort of 256 cognitively normal older people (mean age 55 years) using neuropsychological tests and DNA samples from a study under the direction of Eric Reiman at the Banner Alzheimer’s Institute in Phoenix. Again, KIBRA T allele carriers performed significantly better in two episodic memory tasks, the Buschke’s Selective Reminding Test and the Rey Auditory Verbal Learning Test. But the CLSTN2 SNP failed to show any association with memory, suggesting that it has no effect in the American population or that the effect may be age dependent. In a second Swiss cohort they also found that T allele carriers performed better in tests of visual episodic memory.
What role does KIBRA play in memory and how is that influenced by the intron allele? The authors found that non-T allele carriers exhibited significantly higher neuronal activation when trying to match previously memorized faces and professions. The greater neuronal activation was limited to the hippocampus and parahippocampal gyrus, areas of the brain that are integral to learning and memory, suggesting that the allele affects these areas of the brain in particular (see image below). It appears to have no effect on working memory, executive function, or attention because both carriers and non-carriers of the T allele performed equally well in the Wisconsin Card Sort Test, while in the verbal recall tests there were no differences in immediate recall ability.
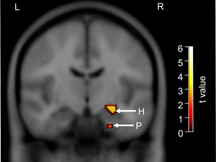
KIBRA Allele Makes a Difference
During a face-profession association task, individuals lacking the KIBRA T allele show significantly greater activation (shown by color-coded t values) in the hippocampus (H) and parahippocampal gyrus (P). The coronal image is made by overlaying data from 15 T allele carriers and 15 non-carriers. [Image © Science]
"The exciting thing about KIBRA is that we completely abandoned the hypothesis-based approach and scanned the genome in an unbiased manner to identify novel genes and pathways related to human memory performance. KIBRA is the first memory-related gene identified by genome-wide scanning. Now that we know of its existence we can track the entire KIBRA-related pathway to understand human memory better and ultimately help people with memory problems,” said Papassotiropoulos.
The authors have examined expression levels of the protein in postmortem human brain and detected the truncated version of the protein (lacking the first 223 amino acids) in memory-related areas, including the hippocampus and temporal lobe. The truncated protein does contain the motifs that interact with dendrin and PKMζ. It will be interesting to see how the SNP might alter expression of the protein or otherwise influence recall.—Tom Fagan.
Q&A with Eric Reiman, Andreas Papassotiropoulos, Dominique de Quervain. and Dietrich Stephan. Questions by Tom Fagan.
Q: How does this data fit with current theories on the effects of genetic variation on memory?
A: Twin studies suggest that about half of the individual differences in normal episodic memory are related to inherited genes. Our studies suggest that the KIBRA gene accounts for about a 20 percent difference between people with better or worse memory performance, remaining significant after correction for more than 500,000 comparisons, and independently confirmed in several independent studies. Each of the studies looked at a slightly different version of episodic memory performance, suggesting that it's related to the ability to recall words or pictures, 5 minutes, 30 minutes, or 24 hours later.
Q: Might these alleles impact memory losses later in life?
A: We do not yet know, but are very interested in determining the extent to which the effects of KIBRA on memory are modified in response to normal aging. While our studies found a difference between KIBRA T carriers and non-carriers in both young and older adults, we would need the same memory test in both groups to determine if KIBRA preferentially affects the non-disabling episodic memory declines that accompany normal aging.
Q: How might these allelic variations relate to mild cognitive impairment, AD, or other dementias? Would you expect those with the T allele to maintain cognition a bit better than those without, for example?
A: Based on our preliminary analyses, the KIBRA gene does not appear to distinguish AD cases from controls—but you ask a very interesting question! If, indeed, KIBRA T non-carriers have less cognitive reserve (at least in the domain of episodic memory), one might predict that they would have an earlier onset of MCI and perhaps dementia in response to the same severity of underlying disease. This is something that could be tested.
Q: How might the KIBRA protein influence human memory?
A: KIBRA's structure suggests at least three possible roles in memory: 1) it may be a modulator of synaptic plasticity, 2) it may interact with the isoform of protein kinase C (PKMζ) that has been implicated in memory formation and long-term potentiation, and 3) it may serve as a calcium sensor in vesicle exocytosis, bolstering the communication between brain cells. In other words, its structure suggests that it could strengthen the connections between brain cells involved in episodic memory by building new connections between these brain cells, by bolstering the electrical response of brain cells to chemical messengers, or by bolstering the transmission of the chemical message itself. Yet it might turn out that KIBRA possesses unique properties unrelated to the above-mentioned mechanisms. It will be very interesting to find out.
Q: Memory-linked genetic variations have been found in genes for BDNF, HTRA2, and now KIBRA. Is there some common denominator or pathway that is emerging?
A: Episodic memory is a highly polygenic trait. While it is possible to identify statistically genetic clusters (de Quervain et al., 2006 and ARF related news story) related to human memory, it is highly premature to interpret these relations in a mechanistical way. The common denominator will be synaptic plasticity after all, which represents an extremely complicated cascade of molecular events (Kandel, 2001).
Q: Does the magnitude of the effect of the BDNF, HRTA2, and KIBRA genetic variations tell us anything about the relative importance of these three proteins in learning and memory?
A: No. The effect of the genetic variations tells us that these genes are important for learning and memory; however, it is not possible to extrapolate a relative contribution. It’s like Alzheimer disease: genetic variations within APP and the presenilins have (if any) a barely measurable effect on AD risk. However, the importance of these genes for the AD-related pathophysiology is huge.
Q: Might the discovery of the KIBRA alleles open any therapeutic avenues for dealing with memory loss or cognitive decline?
A: We're excited about the chance to use this information to identify promising memory-enhancing compounds, several of which we've begun to test in aged rats. The KIBRA protein and its interactions with other proteins permit us to postulate a pathway that may participate in episodic memory. We can consider which compounds affect the actions of these proteins and then test the effects of these potential memory-enhancing compounds on memory performance in certain laboratory animals. The ideal memory-enhancing compound would be effective, extremely safe, and extremely well tolerated. And it could potentially enhance memory performance (even without slowing Alzheimer disease progression) in patients with dementia and MCI, aged persons, and even the public at large, depending on its efficacy, safety, and tolerability.
References
News Citations
- Immune Receptor Controls Synaptic Plasticity; LTP Makes Memories
- Total Recall—Age, Genetic Variation, and Memory
Paper Citations
- Pastalkova E, Serrano P, Pinkhasova D, Wallace E, Fenton AA, Sacktor TC. Storage of spatial information by the maintenance mechanism of LTP. Science. 2006 Aug 25;313(5790):1141-4. PubMed.
- Kremerskothen J, Plaas C, Büther K, Finger I, Veltel S, Matanis T, Liedtke T, Barnekow A. Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun. 2003 Jan 24;300(4):862-7. PubMed.
- de Quervain DJ, Papassotiropoulos A. Identification of a genetic cluster influencing memory performance and hippocampal activity in humans. Proc Natl Acad Sci U S A. 2006 Mar 14;103(11):4270-4. PubMed.
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001 Nov 2;294(5544):1030-8. PubMed.
Other Citations
Further Reading
No Available Further Reading
Primary Papers
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, Wollmer MA, Aerni A, Coluccia D, Hänggi J, Mondadori CR, Buchmann A, Reiman EM, Caselli RJ, Henke K, de Quervain DJ. Common Kibra alleles are associated with human memory performance. Science. 2006 Oct 20;314(5798):475-8. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
SUNY Downstate Medical Center
This paper shows that allelic differences in KIBRA, a protein specifically binding the brain-specific PKC isoform, PKMζ, contribute to variation in human episodic memory performance. This paper follows on the heels of another paper in Science, from our lab, demonstrating that PKMζ is critical for maintaining the hippocampal memory trace in rats for up to one month. The only known function of KIBRA is to bind both PKMζ and the postsynaptic scaffolding protein dendrin; therefore, the new paper suggests that KIBRA is a key player in the localization of PKMζ to synapses. Because mislocalization of PKMζ to the tangles of AD is remarkably specific to tangles in the limbic system (hippocampus, entorhinal cortex, and amygdala) that is essential for episodic memory, and because PKMζ is found only in tangles of people with dementia, and not in the tangles of elderly people without dementia, these new results suggest that the KIBRA-PKMζ pathway is not only an essential pathway for normal memory function, but is the pathway ultimately disrupted in AD that causes loss of memory.
References:
Pastalkova E, Serrano P, Pinkhasova D, Wallace E, Fenton AA, Sacktor TC. Storage of spatial information by the maintenance mechanism of LTP. Science. 2006 Aug 25;313(5790):1141-4. PubMed.
Crary JF, Shao CY, Mirra SS, Hernandez AI, Sacktor TC. Atypical protein kinase C in neurodegenerative disease I: PKMzeta aggregates with limbic neurofibrillary tangles and AMPA receptors in Alzheimer disease. J Neuropathol Exp Neurol. 2006 Apr;65(4):319-26. PubMed.
�
A recent study by Lauriat and colleagues (1) identifies KIBRA, HAX-1 (antiapoptotic mitochondrial protein) and INTS4 as KIAA0513 interacting proteins.
INTS4 is part of the integrator complex that directly interacts with the C-terminal domain of the RNA polymerase II largest subunit (2). It has been reported that long-term memory requires de novo RNA synthesis, and I wonder whether a KIBRA/INTS4 association may also underlie its function in enhancing memory. It is interesting that hyperphosphorylation of RNA polymerase II precedes neurofibrillary tangles in AD (3). Casein kinase 2 phosphorylates RNAP II and a recent study finds that ellagic acid is a potent CK2 inhibitor (4). Does the fact that pomegranate juice contains ellagic acid underlie its benefit in reducing amyloid load? (5) It has also been found to inhibit BACE1 (6).
In his comment, Todd Sacktor suggests that these new results indicate that the KIBRA-PKMζ pathway is not only an essential pathway for normal memory function, but is the pathway ultimately disrupted in AD that causes loss of memory. I also wonder where CAMKII comes into this.
Hongpaisan and colleagues (7) find calcium entering the mitochondria results in increased O2- production that is essential for the activation of CaMKII. They also report that enhanced O2- production also promoted PKC activity.
Studies find reduced pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase activity in AD (8) and reduced ability to sequester cytosolic Ca2+ into mitochondria in pyruvate dehydrogenase deficiency (9). Is there a role for reduced mitochondrial Ca2+ and increased SOD1 which may result in reduced CAMKII and PKC activity in the memory deficits in AD and DS?
Sheline and Mei (10) find that copper facilitated the formation of reactive oxygen species, and inhibited pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase. Do you see altered activity of these enzymes with clioquinol treatment?
Amada and colleagues (11) suggest that N-methyl-D-aspartate receptor deficiency underlies memory impairment in Alzheimer disease, and that this process likely involves insufficient activation of Ca2+/calmodulin-dependent protein kinase II. That may explain the positive reports with the NMDA receptor agonist D-cycloserine. Perhaps the study by Tan (12) may also explain positive results with methylphenidate. He found that CaM-kinase II in the hippocampus and nucleus accumbens of methylphenidate-injected rats had significantly higher calcium-independent activity compared to the controls at 12 weeks old.
The study by Lindhurst et al. (13) finding that Slc25a19 is a thiamine pyrophosphate transporter is most interesting. Mutations in this gene cause Amish lethal microcephaly. Thiamine pyrophosphate is a cofactor for pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase. Might reduced activity of these enzymes in DS due to SOD1 overexpression also cause microcephaly? Does reduced activity also release the cofactor and explain the increased serum thiamine pyrophosphate in patients who have DS? Could we expect benefit with thiamine pyrophosphate supplementation?
References:
Lauriat TL, Dracheva S, Kremerskothen J, Duning K, Haroutunian V, Buxbaum JD, Hyde TM, Kleinman JE, McInnes LA. Characterization of KIAA0513, a novel signaling molecule that interacts with modulators of neuroplasticity, apoptosis, and the cytoskeleton. Brain Res. 2006 Nov 22;1121(1):1-11. PubMed.
Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005 Oct 21;123(2):265-76. PubMed.
Husseman JW, Hallows JL, Bregman DB, Leverenz JB, Nochlin D, Jin LW, Vincent I. Hyperphosphorylation of RNA polymerase II and reduced neuronal RNA levels precede neurofibrillary tangles in Alzheimer disease. J Neuropathol Exp Neurol. 2001 Dec;60(12):1219-32. PubMed.
Cozza G, Bonvini P, Zorzi E, Poletto G, Pagano MA, Sarno S, Donella-Deana A, Zagotto G, Rosolen A, Pinna LA, Meggio F, Moro S. Identification of ellagic acid as potent inhibitor of protein kinase CK2: a successful example of a virtual screening application. J Med Chem. 2006 Apr 20;49(8):2363-6. PubMed.
Hartman RE, Shah A, Fagan AM, Schwetye KE, Parsadanian M, Schulman RN, Finn MB, Holtzman DM. Pomegranate juice decreases amyloid load and improves behavior in a mouse model of Alzheimer's disease. Neurobiol Dis. 2006 Dec;24(3):506-15. PubMed.
Kwak HM, Jeon SY, Sohng BH, Kim JG, Lee JM, Lee KB, Jeong HH, Hur JM, Kang YH, Song KS. beta-Secretase (BACE1) inhibitors from pomegranate (Punica granatum) husk. Arch Pharm Res. 2005 Dec;28(12):1328-32. PubMed.
Hongpaisan J, Winters CA, Andrews SB. Strong calcium entry activates mitochondrial superoxide generation, upregulating kinase signaling in hippocampal neurons. J Neurosci. 2004 Dec 1;24(48):10878-87. PubMed.
Bubber P, Haroutunian V, Fisch G, Blass JP, Gibson GE. Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann Neurol. 2005 May;57(5):695-703. PubMed.
Padua RA, Baron KT, Thyagarajan B, Campbell C, Thayer SA. Reduced Ca2+ uptake by mitochondria in pyruvate dehydrogenase-deficient human diploid fibroblasts. Am J Physiol. 1998 Mar;274(3 Pt 1):C615-22. PubMed.
Sheline CT, Wei L. Free radical-mediated neurotoxicity may be caused by inhibition of mitochondrial dehydrogenases in vitro and in vivo. Neuroscience. 2006 Jun 19;140(1):235-46. PubMed.
Amada N, Aihara K, Ravid R, Horie M. Reduction of NR1 and phosphorylated Ca2+/calmodulin-dependent protein kinase II levels in Alzheimer's disease. Neuroreport. 2005 Nov 7;16(16):1809-13. PubMed.
Tan SE. The activations of calcium/calmodulin-dependent protein kinase II in hippocampus and nucleus accumbens of rats after methylphenidate injection. Kaohsiung J Med Sci. 1999 Oct;15(10):625-31. PubMed.
Lindhurst MJ, Fiermonte G, Song S, Struys E, De Leonardis F, Schwartzberg PL, Chen A, Castegna A, Verhoeven N, Mathews CK, Palmieri F, Biesecker LG. Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc Natl Acad Sci U S A. 2006 Oct 24;103(43):15927-32. PubMed.
Make a Comment
To make a comment you must login or register.