Quantum Leap? Nanoprobes Track Aβ Aggregation in Real Time
Quick Links
With newly developed quantum-dot nanoprobes, scientists can now watch individual Aβ molecules glom together into oligomers and fibrils in real time. The technology may find use in high-throughput drug screening since it is quantitative, uses tiny samples, and detects various forms of Aβ aggregates. Senior investigator Tsuneya Ikezu of the University of Nebraska, Omaha, and colleagues reported the work in the December issue of PLoS ONE.
Technological advancement has given researchers a growing number of tools for visualizing amyloid-β peptides, including two-photon microscopy for real-time measurement of plaque growth in AD mouse models (Yan et al., 2009 and Meyer-Luehmann et al., 2008). However, Ikezu’s study is the first to dynamically and quantitatively track formation of amyloid-β oligomers, now widely seen as the most neurotoxic Aβ species. Last year, researchers published an enzyme-linked immunosorbent assay (ELISA) specific for Aβ oligomers (Xia et al., 2009 and ARF related news story), but when it comes to live imaging, standard reagents for plaque staining, e.g., thioflavin or Congo red, bind poorly to the β-sheet structure of Aβ oligomers. They also poorly penetrate the brain. Ikezu’s lab found several β-sheet binding fluorescent dyes that do reach the brain, e.g., X-34, FSB, and derivatives (Flaherty et al., 2007), but all these amyloid imaging compounds have a key problem. “Once they bind to the plaque, they inhibit aggregation,” Ikezu said. “They competitively interfere with assembly of the β-sheet structure.” Furthermore, the short half-life of these reagents precludes their use in long-term imaging studies.
Quantum dots (QD), on the other hand, are fluorescent semiconductor nanocrystals that “emit signals for a long time, almost indefinitely,” Ikezu said. “You keep [the QD probe] on ice for one month, and it’s still usable.” A previous report described Aβ imaging using QD, but the labeling relied on non-specific ionic interactions between QD particles and Aβ fibrils (Ji et al., 2006). Ikezu’s team, led by first author Kiyotaka Tokuraku, is the first to make Aβ-specific nanoprobes by directly coupling Aβ peptide to QDs. Tokuraku, a professor at Miyakonojo National College of Technology, Japan, did the work during his recent sabbatical in the Ikezu lab.
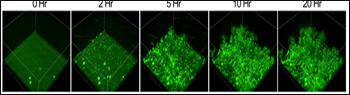
Aggregation in Real Time
Quantum-dot Aβ nanoprobes (green) were incubated with free Aβ42 (to enable aggregation) on a 96-well plate and visualized by confocal microscopy every 30 minutes for 20 hours. To see a dynamic rendition of Aβ aggregation, see movie (the movie takes a few seconds to download). Additional movies are also available as supporting information in the paper. Image credit: Tsuneya Ikezu and PLoS ONE
The researchers attached a crosslinker to the QDs before coupling them to Aβ40 peptides. They used QDAβ to measure Aβ oligomerization in vitro, showing that the size of Aβ aggregates could be estimated from relative intensities of spots imaged by fluorescence microscopy. The QD probes could also monitor Aβ fibrillization (see image), as long as non-conjugated Aβ was mixed with the QDAβ. (Because QD particles are quite large relative to Aβ peptides, it is likely that QD steric hindrance inhibits fibril formation.)
Ikezu suggested that the Aβ QD nanoprobes would be good for single-molecule imaging of Aβ in cultured cells—seeing how Aβ oligomers get endocytosed, for example. As described in the current report, his team has used QDAβ to watch microglia gobble up Aβ. The phagocytes seemed to prefer monomeric QDAβ over oligomers, and the ingested QDAβ mainly accumulated in the lysosome.
Another potential application for QDAβ is small-scale drug screening. “You can see how drugs block different kinds of Aβ aggregates,” Ikezu said. He and colleagues showed that Aβ-specific antibodies, but not control antibodies to tubulin, were able to block QDAβ aggregation.—Esther Landhuis
References
News Citations
Paper Citations
- Yan P, Bero AW, Cirrito JR, Xiao Q, Hu X, Wang Y, Gonzales E, Holtzman DM, Lee JM. Characterizing the appearance and growth of amyloid plaques in APP/PS1 mice. J Neurosci. 2009 Aug 26;29(34):10706-14. PubMed.
- Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer's disease. Nature. 2008 Feb 7;451(7179):720-4. PubMed.
- Xia W, Yang T, Shankar G, Smith IM, Shen Y, Walsh DM, Selkoe DJ. A specific enzyme-linked immunosorbent assay for measuring beta-amyloid protein oligomers in human plasma and brain tissue of patients with Alzheimer disease. Arch Neurol. 2009 Feb;66(2):190-9. PubMed.
- Flaherty DP, Walsh SM, Kiyota T, Dong Y, Ikezu T, Vennerstrom JL. Polyfluorinated bis-styrylbenzene beta-amyloid plaque binding ligands. J Med Chem. 2007 Oct 4;50(20):4986-92. PubMed.
- Ji X, Naistat D, Li C, Orbulesco J, Leblanc RM. An alternative approach to amyloid fibrils morphology: CdSe/ZnS quantum dots labelled beta-amyloid peptide fragments Abeta (31-35), Abeta (1-40) and Abeta (1-42). Colloids Surf B Biointerfaces. 2006 Jul 1;50(2):104-11. PubMed.
External Citations
Further Reading
Primary Papers
- Tokuraku K, Marquardt M, Ikezu T. Real-time imaging and quantification of amyloid-beta peptide aggregates by novel quantum-dot nanoprobes. PLoS One. 2009;4(12):e8492. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.